Evelina Ametova
Shot Noise Reduction in Radiographic and Tomographic Multi-Channel Imaging with Self-Supervised Deep Learning
Mar 25, 2023Abstract:Noise is an important issue for radiographic and tomographic imaging techniques. It becomes particularly critical in applications where additional constraints force a strong reduction of the Signal-to-Noise Ratio (SNR) per image. These constraints may result from limitations on the maximum available flux or permissible dose and the associated restriction on exposure time. Often, a high SNR per image is traded for the ability to distribute a given total exposure capacity per pixel over multiple channels, thus obtaining additional information about the object by the same total exposure time. These can be energy channels in the case of spectroscopic imaging or time channels in the case of time-resolved imaging. In this paper, we report on a method for improving the quality of noisy multi-channel (time or energy-resolved) imaging datasets. The method relies on the recent Noise2Noise (N2N) self-supervised denoising approach that learns to predict a noise-free signal without access to noise-free data. N2N in turn requires drawing pairs of samples from a data distribution sharing identical signals while being exposed to different samples of random noise. The method is applicable if adjacent channels share enough information to provide images with similar enough information but independent noise. We demonstrate several representative case studies, namely spectroscopic (k-edge) X-ray tomography, in vivo X-ray cine-radiography, and energy-dispersive (Bragg edge) neutron tomography. In all cases, the N2N method shows dramatic improvement and outperforms conventional denoising methods. For such imaging techniques, the method can therefore significantly improve image quality, or maintain image quality with further reduced exposure time per image.
Enhanced hyperspectral tomography for bioimaging by spatiospectral reconstruction
Mar 08, 2021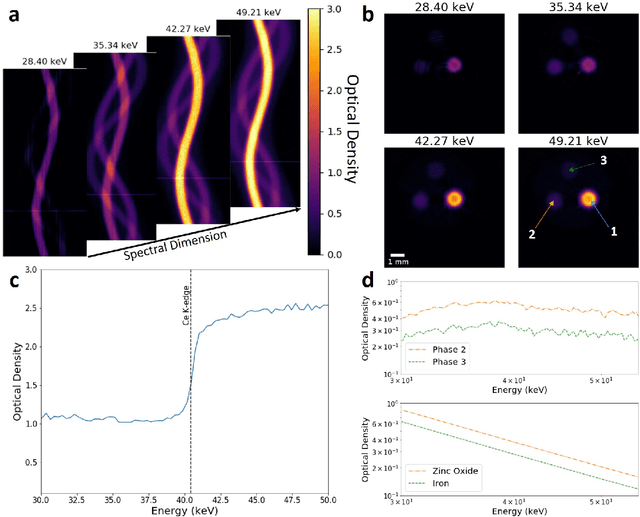
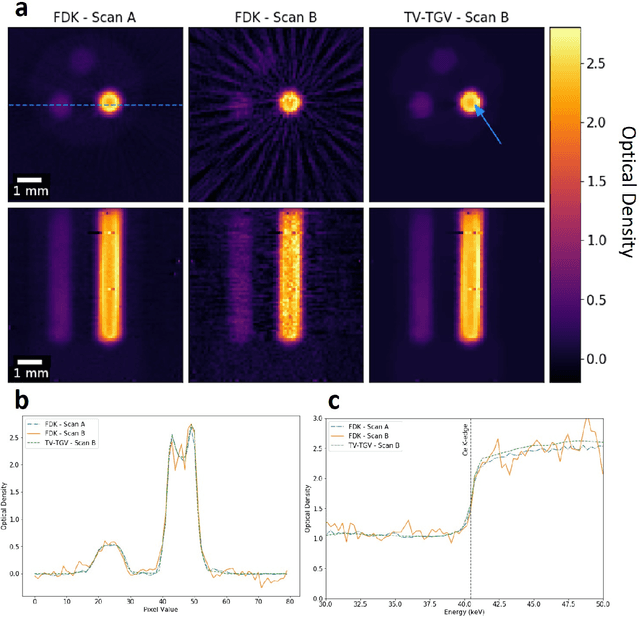
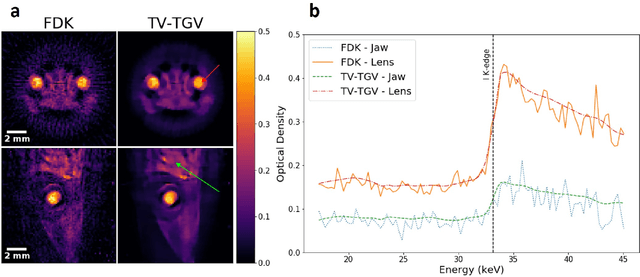
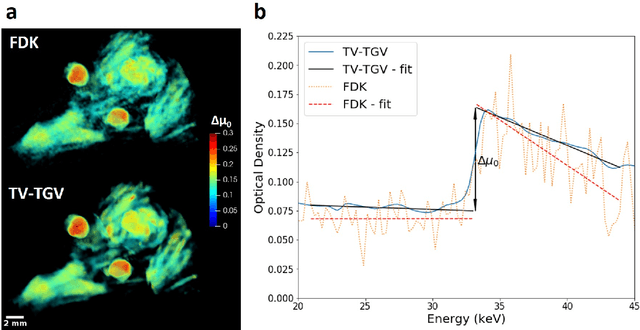
Abstract:Here we apply hyperspectral bright field imaging to collect computed tomographic images with excellent energy resolution (800 eV), applying it for the first time to map the distribution of stain in a fixed biological sample through its characteristic K-edge. Conventionally, because the photons detected at each pixel are distributed across as many as 200 energy channels, energy-selective images are characterised by low count-rates and poor signal-to-noise ratio. This means high X-ray exposures, long scan times and high doses are required to image unique spectral markers. Here, we achieve high quality energy-dispersive tomograms from low dose, noisy datasets using a dedicated iterative reconstruction algorithm. This exploits the spatial smoothness and inter-channel structural correlation in the spectral domain using two carefully chosen regularisation terms. For a multi-phase phantom, a reduction in scan time of 36 times is demonstrated. Spectral analysis methods including K-edge subtraction and absorption step-size fitting are evaluated for an ex vivo, single (iodine)-stained biological sample, where low chemical concentration and inhomogeneous distribution can affect soft tissue segmentation and visualisation. The reconstruction algorithms are available through the open-source Core Imaging Library. Taken together, these tools offer new capabilities for visualisation and elemental mapping, with promising applications for multiply-stained biological specimens.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge