Milan Sulc
FungiTastic: A multi-modal dataset and benchmark for image categorization
Aug 24, 2024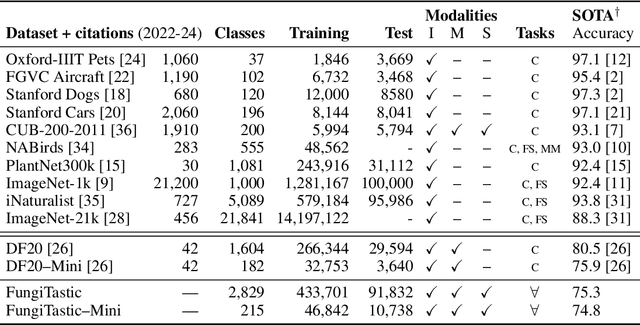
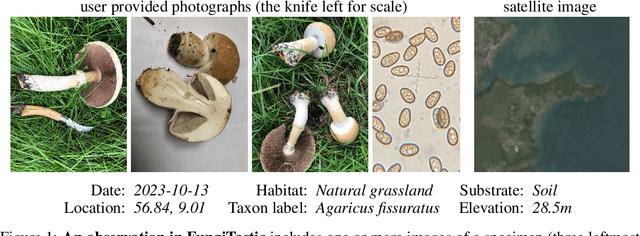
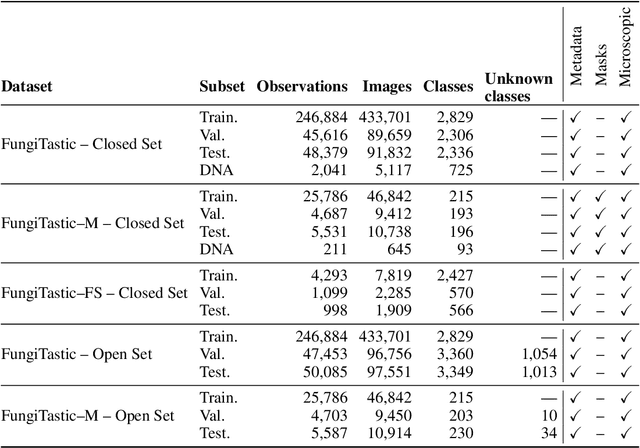
Abstract:We introduce a new, highly challenging benchmark and a dataset -- FungiTastic -- based on data continuously collected over a twenty-year span. The dataset originates in fungal records labeled and curated by experts. It consists of about 350k multi-modal observations that include more than 650k photographs from 5k fine-grained categories and diverse accompanying information, e.g., acquisition metadata, satellite images, and body part segmentation. FungiTastic is the only benchmark that includes a test set with partially DNA-sequenced ground truth of unprecedented label reliability. The benchmark is designed to support (i) standard close-set classification, (ii) open-set classification, (iii) multi-modal classification, (iv) few-shot learning, (v) domain shift, and many more. We provide baseline methods tailored for almost all the use-cases. We provide a multitude of ready-to-use pre-trained models on HuggingFace and a framework for model training. A comprehensive documentation describing the dataset features and the baselines are available at https://bohemianvra.github.io/FungiTastic/ and https://www.kaggle.com/datasets/picekl/fungitastic.
Contrastive Classification and Representation Learning with Probabilistic Interpretation
Nov 07, 2022



Abstract:Cross entropy loss has served as the main objective function for classification-based tasks. Widely deployed for learning neural network classifiers, it shows both effectiveness and a probabilistic interpretation. Recently, after the success of self supervised contrastive representation learning methods, supervised contrastive methods have been proposed to learn representations and have shown superior and more robust performance, compared to solely training with cross entropy loss. However, cross entropy loss is still needed to train the final classification layer. In this work, we investigate the possibility of learning both the representation and the classifier using one objective function that combines the robustness of contrastive learning and the probabilistic interpretation of cross entropy loss. First, we revisit a previously proposed contrastive-based objective function that approximates cross entropy loss and present a simple extension to learn the classifier jointly. Second, we propose a new version of the supervised contrastive training that learns jointly the parameters of the classifier and the backbone of the network. We empirically show that our proposed objective functions show a significant improvement over the standard cross entropy loss with more training stability and robustness in various challenging settings.
The Hitchhiker's Guide to Prior-Shift Adaptation
Jun 22, 2021



Abstract:In many computer vision classification tasks, class priors at test time often differ from priors on the training set. In the case of such prior shift, classifiers must be adapted correspondingly to maintain close to optimal performance. This paper analyzes methods for adaptation of probabilistic classifiers to new priors and for estimating new priors on an unlabeled test set. We propose a novel method to address a known issue of prior estimation methods based on confusion matrices, where inconsistent estimates of decision probabilities and confusion matrices lead to negative values in the estimated priors. Experiments on fine-grained image classification datasets provide insight into the best practice of prior shift estimation and classifier adaptation and show that the proposed method achieves state-of-the-art results in prior adaptation. Applying the best practice to two tasks with naturally imbalanced priors, learning from web-crawled images and plant species classification, increased the recognition accuracy by 1.1% and 3.4% respectively.
Improving CNN classifiers by estimating test-time priors
May 21, 2018



Abstract:The problem of different training and test set class priors is addressed in the context of CNN classifiers. An EM-based algorithm for test-time class priors estimation is evaluated on fine-grained computer vision problems for both the batch and on-line situations. Experimental results show a significant improvement on the fine-grained classification tasks using the known evaluation-time priors, increasing the top-1 accuracy by 4.0% on the FGVC iNaturalist 2018 validation set and by 3.9% on the FGVCx Fungi 2018 validation set. Iterative estimation of test-time priors on the PlantCLEF 2017 dataset increased the image classification accuracy by 3.4%, allowing a single CNN model to achieve state-of-the-art results and outperform the competition-winning ensemble of 12 CNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge