Lucy Fortson
Scaling Laws for Galaxy Images
Apr 03, 2024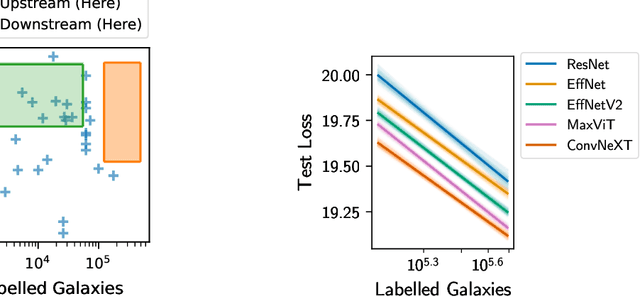
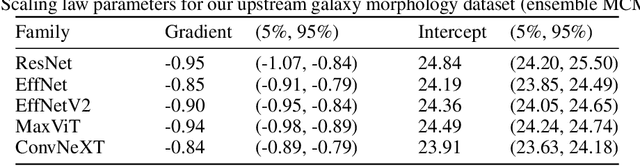
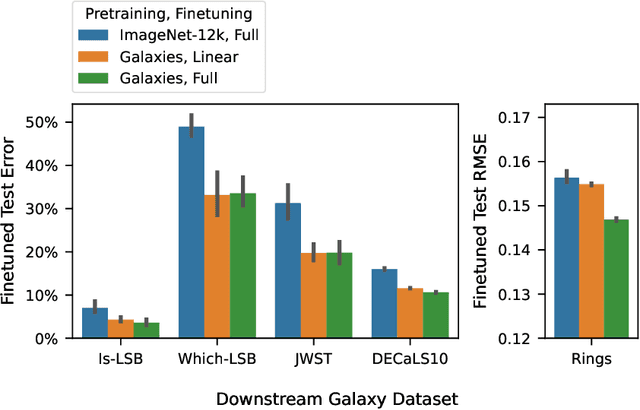
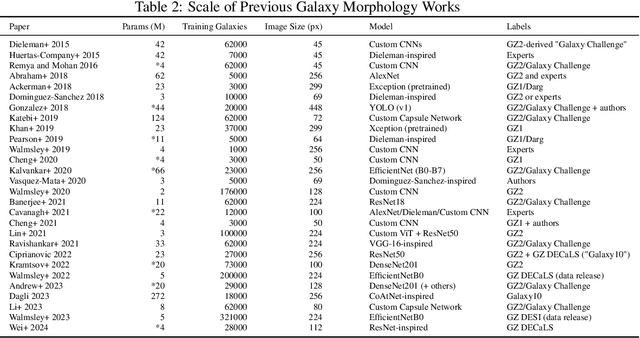
Abstract:We present the first systematic investigation of supervised scaling laws outside of an ImageNet-like context - on images of galaxies. We use 840k galaxy images and over 100M annotations by Galaxy Zoo volunteers, comparable in scale to Imagenet-1K. We find that adding annotated galaxy images provides a power law improvement in performance across all architectures and all tasks, while adding trainable parameters is effective only for some (typically more subjectively challenging) tasks. We then compare the downstream performance of finetuned models pretrained on either ImageNet-12k alone vs. additionally pretrained on our galaxy images. We achieve an average relative error rate reduction of 31% across 5 downstream tasks of scientific interest. Our finetuned models are more label-efficient and, unlike their ImageNet-12k-pretrained equivalents, often achieve linear transfer performance equal to that of end-to-end finetuning. We find relatively modest additional downstream benefits from scaling model size, implying that scaling alone is not sufficient to address our domain gap, and suggest that practitioners with qualitatively different images might benefit more from in-domain adaption followed by targeted downstream labelling.
Automated 3D Tumor Segmentation using Temporal Cubic PatchGAN (TCuP-GAN)
Nov 23, 2023Abstract:Development of robust general purpose 3D segmentation frameworks using the latest deep learning techniques is one of the active topics in various bio-medical domains. In this work, we introduce Temporal Cubic PatchGAN (TCuP-GAN), a volume-to-volume translational model that marries the concepts of a generative feature learning framework with Convolutional Long Short-Term Memory Networks (LSTMs), for the task of 3D segmentation. We demonstrate the capabilities of our TCuP-GAN on the data from four segmentation challenges (Adult Glioma, Meningioma, Pediatric Tumors, and Sub-Saharan Africa subset) featured within the 2023 Brain Tumor Segmentation (BraTS) Challenge and quantify its performance using LesionWise Dice similarity and $95\%$ Hausdorff Distance metrics. We demonstrate the successful learning of our framework to predict robust multi-class segmentation masks across all the challenges. This benchmarking work serves as a stepping stone for future efforts towards applying TCuP-GAN on other multi-class tasks such as multi-organelle segmentation in electron microscopy imaging.
TCuPGAN: A novel framework developed for optimizing human-machine interactions in citizen science
Nov 23, 2023Abstract:In the era of big data in scientific research, there is a necessity to leverage techniques which reduce human effort in labeling and categorizing large datasets by involving sophisticated machine tools. To combat this problem, we present a novel, general purpose model for 3D segmentation that leverages patch-wise adversariality and Long Short-Term Memory to encode sequential information. Using this model alongside citizen science projects which use 3D datasets (image cubes) on the Zooniverse platforms, we propose an iterative human-machine optimization framework where only a fraction of the 2D slices from these cubes are seen by the volunteers. We leverage the patch-wise discriminator in our model to provide an estimate of which slices within these image cubes have poorly generalized feature representations, and correspondingly poor machine performance. These images with corresponding machine proposals would be presented to volunteers on Zooniverse for correction, leading to a drastic reduction in the volunteer effort on citizen science projects. We trained our model on ~2300 liver tissue 3D electron micrographs. Lipid droplets were segmented within these images through human annotation via the `Etch A Cell - Fat Checker' citizen science project, hosted on the Zooniverse platform. In this work, we demonstrate this framework and the selection methodology which resulted in a measured reduction in volunteer effort by more than 60%. We envision this type of joint human-machine partnership will be of great use on future Zooniverse projects.
From fat droplets to floating forests: cross-domain transfer learning using a PatchGAN-based segmentation model
Nov 08, 2022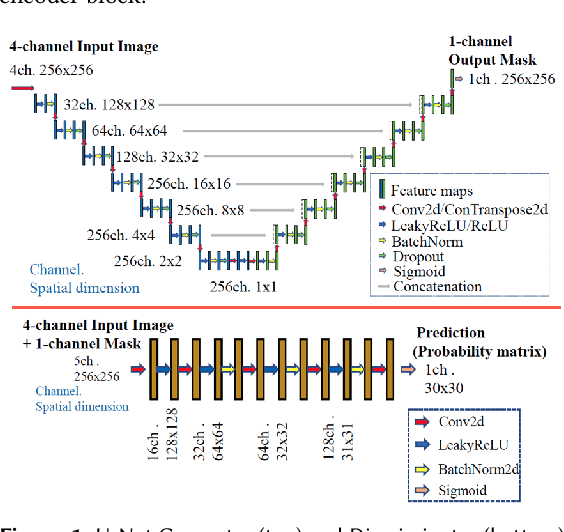
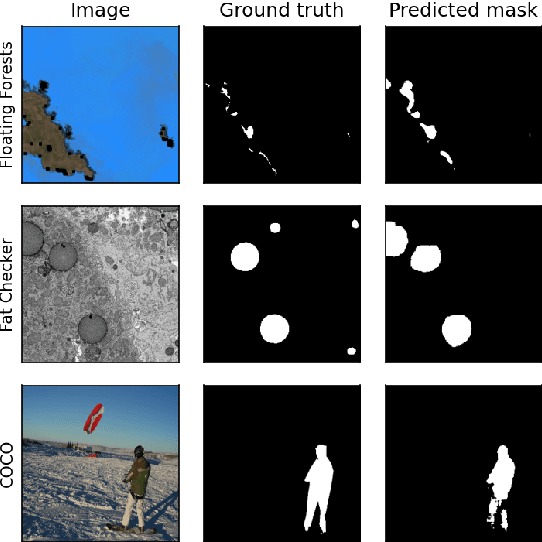
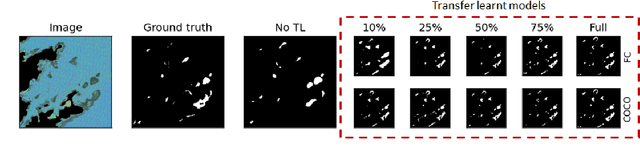
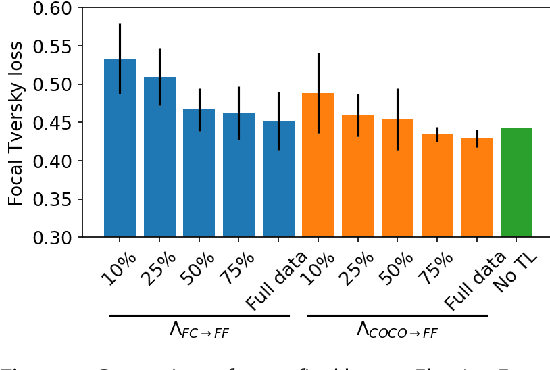
Abstract:Many scientific domains gather sufficient labels to train machine algorithms through human-in-the-loop techniques provided by the Zooniverse.org citizen science platform. As the range of projects, task types and data rates increase, acceleration of model training is of paramount concern to focus volunteer effort where most needed. The application of Transfer Learning (TL) between Zooniverse projects holds promise as a solution. However, understanding the effectiveness of TL approaches that pretrain on large-scale generic image sets vs. images with similar characteristics possibly from similar tasks is an open challenge. We apply a generative segmentation model on two Zooniverse project-based data sets: (1) to identify fat droplets in liver cells (FatChecker; FC) and (2) the identification of kelp beds in satellite images (Floating Forests; FF) through transfer learning from the first project. We compare and contrast its performance with a TL model based on the COCO image set, and subsequently with baseline counterparts. We find that both the FC and COCO TL models perform better than the baseline cases when using >75% of the original training sample size. The COCO-based TL model generally performs better than the FC-based one, likely due to its generalized features. Our investigations provide important insights into usage of TL approaches on multi-domain data hosted across different Zooniverse projects, enabling future projects to accelerate task completion.
Practical Galaxy Morphology Tools from Deep Supervised Representation Learning
Oct 25, 2021
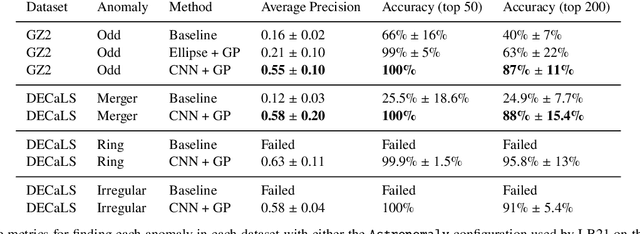
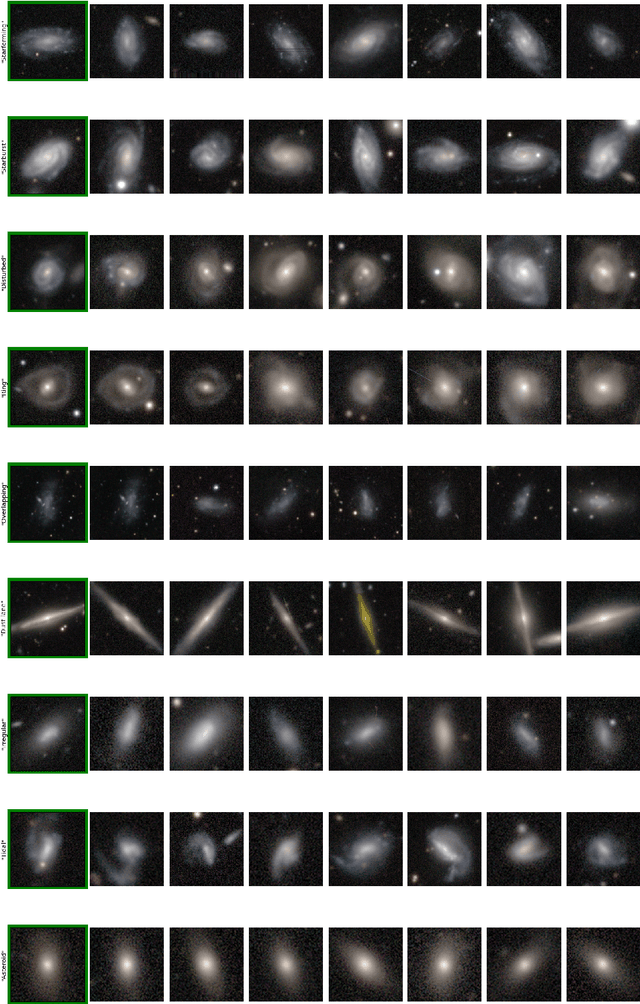
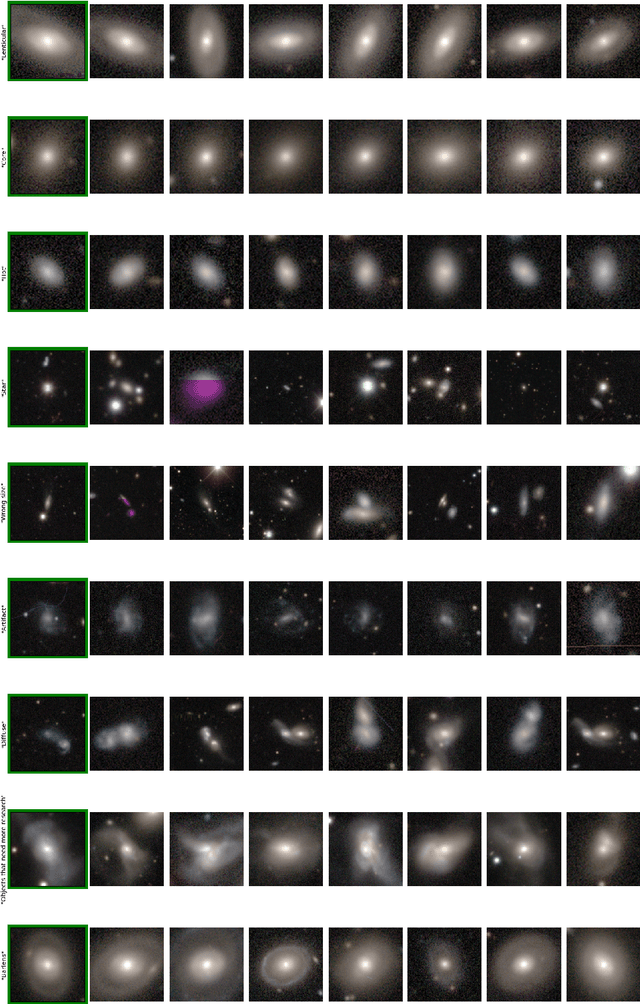
Abstract:Astronomers have typically set out to solve supervised machine learning problems by creating their own representations from scratch. We show that deep learning models trained to answer every Galaxy Zoo DECaLS question learn meaningful semantic representations of galaxies that are useful for new tasks on which the models were never trained. We exploit these representations to outperform existing approaches at several practical tasks crucial for investigating large galaxy samples. The first task is identifying galaxies of similar morphology to a query galaxy. Given a single galaxy assigned a free text tag by humans (e.g. `#diffuse'), we can find galaxies matching that tag for most tags. The second task is identifying the most interesting anomalies to a particular researcher. Our approach is 100\% accurate at identifying the most interesting 100 anomalies (as judged by Galaxy Zoo 2 volunteers). The third task is adapting a model to solve a new task using only a small number of newly-labelled galaxies. Models fine-tuned from our representation are better able to identify ring galaxies than models fine-tuned from terrestrial images (ImageNet) or trained from scratch. We solve each task with very few new labels; either one (for the similarity search) or several hundred (for anomaly detection or fine-tuning). This challenges the longstanding view that deep supervised methods require new large labelled datasets for practical use in astronomy. To help the community benefit from our pretrained models, we release our fine-tuning code zoobot. Zoobot is accessible to researchers with no prior experience in deep learning.
Imagine All the People: Citizen Science, Artificial Intelligence, and Computational Research
Apr 05, 2021Abstract:Machine learning, artificial intelligence, and deep learning have advanced significantly over the past decade. Nonetheless, humans possess unique abilities such as creativity, intuition, context and abstraction, analytic problem solving, and detecting unusual events. To successfully tackle pressing scientific and societal challenges, we need the complementary capabilities of both humans and machines. The Federal Government could accelerate its priorities on multiple fronts through judicious integration of citizen science and crowdsourcing with artificial intelligence (AI), Internet of Things (IoT), and cloud strategies.
Galaxy Zoo DECaLS: Detailed Visual Morphology Measurements from Volunteers and Deep Learning for 314,000 Galaxies
Feb 16, 2021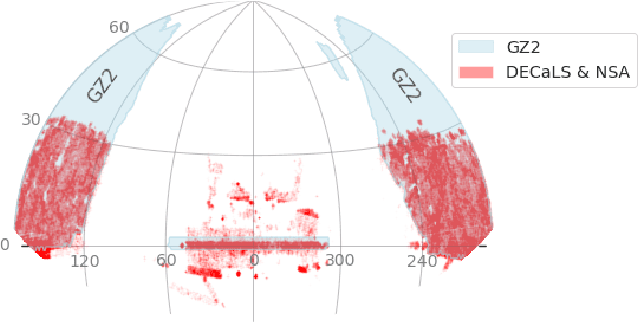
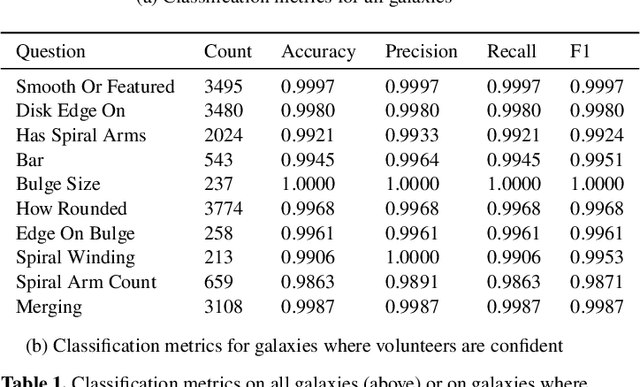
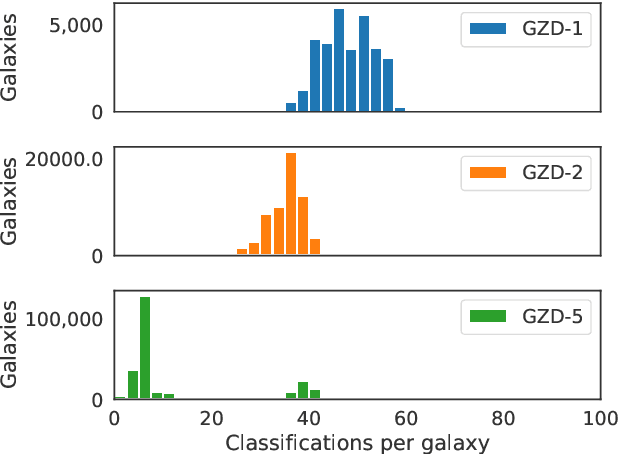
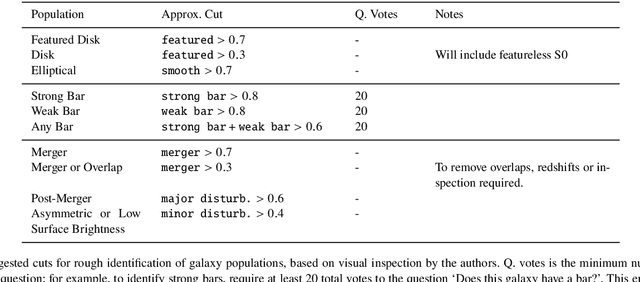
Abstract:We present Galaxy Zoo DECaLS: detailed visual morphological classifications for Dark Energy Camera Legacy Survey images of galaxies within the SDSS DR8 footprint. Deeper DECaLS images (r=23.6 vs. r=22.2 from SDSS) reveal spiral arms, weak bars, and tidal features not previously visible in SDSS imaging. To best exploit the greater depth of DECaLS images, volunteers select from a new set of answers designed to improve our sensitivity to mergers and bars. Galaxy Zoo volunteers provide 7.5 million individual classifications over 314,000 galaxies. 140,000 galaxies receive at least 30 classifications, sufficient to accurately measure detailed morphology like bars, and the remainder receive approximately 5. All classifications are used to train an ensemble of Bayesian convolutional neural networks (a state-of-the-art deep learning method) to predict posteriors for the detailed morphology of all 314,000 galaxies. When measured against confident volunteer classifications, the networks are approximately 99% accurate on every question. Morphology is a fundamental feature of every galaxy; our human and machine classifications are an accurate and detailed resource for understanding how galaxies evolve.
Galaxy Zoo: Probabilistic Morphology through Bayesian CNNs and Active Learning
May 17, 2019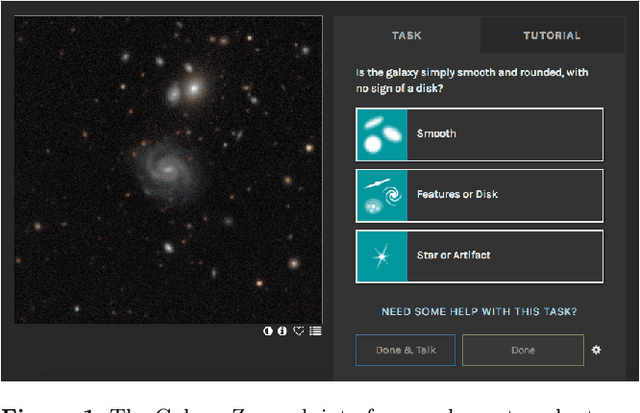

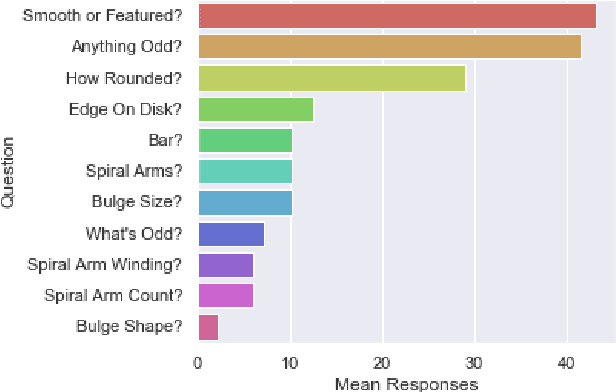

Abstract:We use Bayesian convolutional neural networks and a novel generative model of Galaxy Zoo volunteer responses to infer posteriors for the visual morphology of galaxies. Bayesian CNN can learn from galaxy images with uncertain labels and then, for previously unlabelled galaxies, predict the probability of each possible label. Our posteriors are well-calibrated (e.g. for predicting bars, we achieve coverage errors of 10.6% within 5 responses and 2.9% within 10 responses) and hence are reliable for practical use. Further, using our posteriors, we apply the active learning strategy BALD to request volunteer responses for the subset of galaxies which, if labelled, would be most informative for training our network. We show that training our Bayesian CNNs using active learning requires up to 35-60% fewer labelled galaxies, depending on the morphological feature being classified. By combining human and machine intelligence, Galaxy Zoo will be able to classify surveys of any conceivable scale on a timescale of weeks, providing massive and detailed morphology catalogues to support research into galaxy evolution.
Citizen Science: Contributions to Astronomy Research
Feb 12, 2012
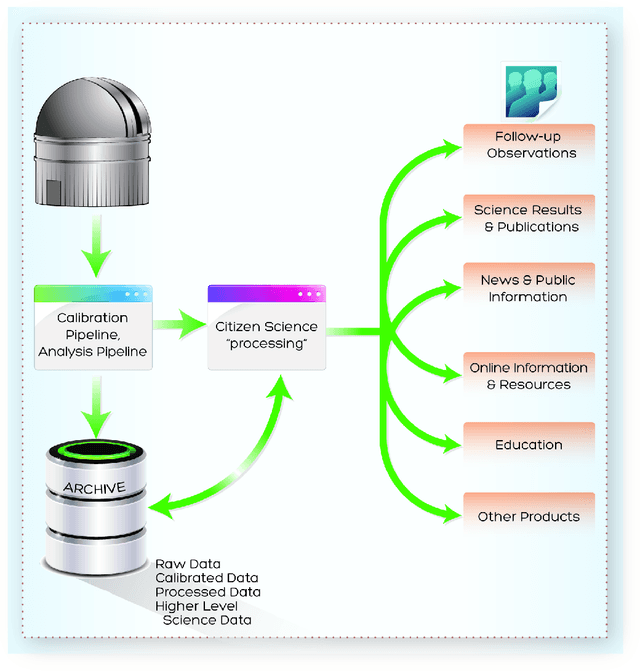
Abstract:The contributions of everyday individuals to significant research has grown dramatically beyond the early days of classical birdwatching and endeavors of amateurs of the 19th century. Now people who are casually interested in science can participate directly in research covering diverse scientific fields. Regarding astronomy, volunteers, either as individuals or as networks of people, are involved in a variety of types of studies. Citizen Science is intuitive, engaging, yet necessarily robust in its adoption of sci-entific principles and methods. Herein, we discuss Citizen Science, focusing on fully participatory projects such as Zooniverse (by several of the au-thors CL, AS, LF, SB), with mention of other programs. In particular, we make the case that citizen science (CS) can be an important aspect of the scientific data analysis pipelines provided to scientists by observatories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge