Kameswara Bharadwaj Mantha
Automated 3D Tumor Segmentation using Temporal Cubic PatchGAN (TCuP-GAN)
Nov 23, 2023Abstract:Development of robust general purpose 3D segmentation frameworks using the latest deep learning techniques is one of the active topics in various bio-medical domains. In this work, we introduce Temporal Cubic PatchGAN (TCuP-GAN), a volume-to-volume translational model that marries the concepts of a generative feature learning framework with Convolutional Long Short-Term Memory Networks (LSTMs), for the task of 3D segmentation. We demonstrate the capabilities of our TCuP-GAN on the data from four segmentation challenges (Adult Glioma, Meningioma, Pediatric Tumors, and Sub-Saharan Africa subset) featured within the 2023 Brain Tumor Segmentation (BraTS) Challenge and quantify its performance using LesionWise Dice similarity and $95\%$ Hausdorff Distance metrics. We demonstrate the successful learning of our framework to predict robust multi-class segmentation masks across all the challenges. This benchmarking work serves as a stepping stone for future efforts towards applying TCuP-GAN on other multi-class tasks such as multi-organelle segmentation in electron microscopy imaging.
From fat droplets to floating forests: cross-domain transfer learning using a PatchGAN-based segmentation model
Nov 08, 2022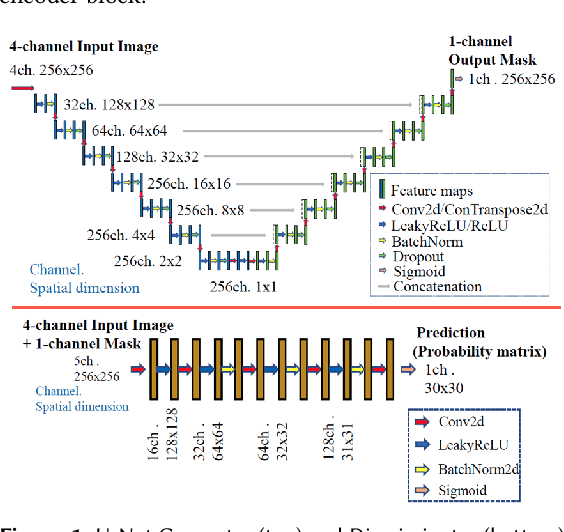
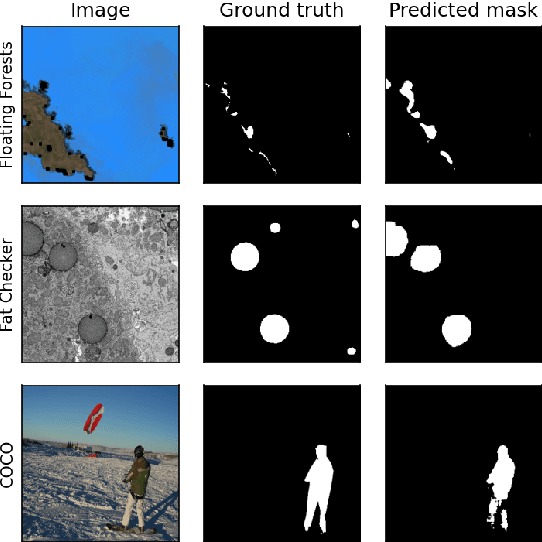
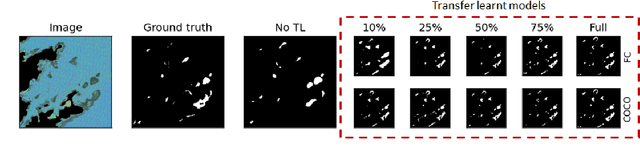
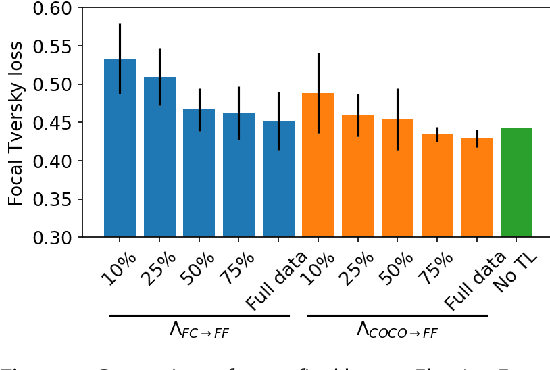
Abstract:Many scientific domains gather sufficient labels to train machine algorithms through human-in-the-loop techniques provided by the Zooniverse.org citizen science platform. As the range of projects, task types and data rates increase, acceleration of model training is of paramount concern to focus volunteer effort where most needed. The application of Transfer Learning (TL) between Zooniverse projects holds promise as a solution. However, understanding the effectiveness of TL approaches that pretrain on large-scale generic image sets vs. images with similar characteristics possibly from similar tasks is an open challenge. We apply a generative segmentation model on two Zooniverse project-based data sets: (1) to identify fat droplets in liver cells (FatChecker; FC) and (2) the identification of kelp beds in satellite images (Floating Forests; FF) through transfer learning from the first project. We compare and contrast its performance with a TL model based on the COCO image set, and subsequently with baseline counterparts. We find that both the FC and COCO TL models perform better than the baseline cases when using >75% of the original training sample size. The COCO-based TL model generally performs better than the FC-based one, likely due to its generalized features. Our investigations provide important insights into usage of TL approaches on multi-domain data hosted across different Zooniverse projects, enabling future projects to accelerate task completion.
Practical Galaxy Morphology Tools from Deep Supervised Representation Learning
Oct 25, 2021
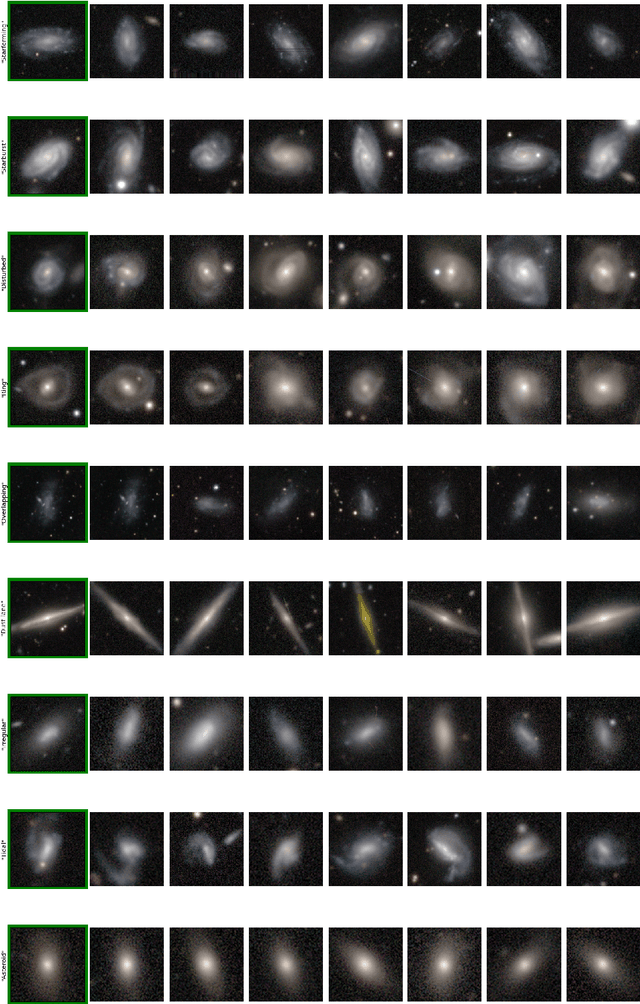
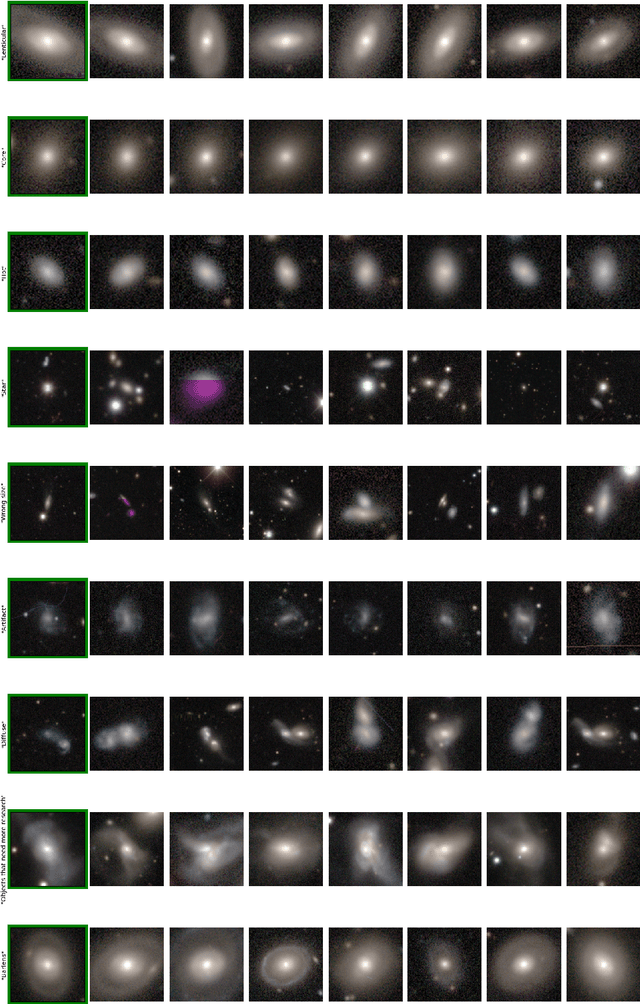
Abstract:Astronomers have typically set out to solve supervised machine learning problems by creating their own representations from scratch. We show that deep learning models trained to answer every Galaxy Zoo DECaLS question learn meaningful semantic representations of galaxies that are useful for new tasks on which the models were never trained. We exploit these representations to outperform existing approaches at several practical tasks crucial for investigating large galaxy samples. The first task is identifying galaxies of similar morphology to a query galaxy. Given a single galaxy assigned a free text tag by humans (e.g. `#diffuse'), we can find galaxies matching that tag for most tags. The second task is identifying the most interesting anomalies to a particular researcher. Our approach is 100\% accurate at identifying the most interesting 100 anomalies (as judged by Galaxy Zoo 2 volunteers). The third task is adapting a model to solve a new task using only a small number of newly-labelled galaxies. Models fine-tuned from our representation are better able to identify ring galaxies than models fine-tuned from terrestrial images (ImageNet) or trained from scratch. We solve each task with very few new labels; either one (for the similarity search) or several hundred (for anomaly detection or fine-tuning). This challenges the longstanding view that deep supervised methods require new large labelled datasets for practical use in astronomy. To help the community benefit from our pretrained models, we release our fine-tuning code zoobot. Zoobot is accessible to researchers with no prior experience in deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge