Hugh Dickinson
Scaling Laws for Galaxy Images
Apr 03, 2024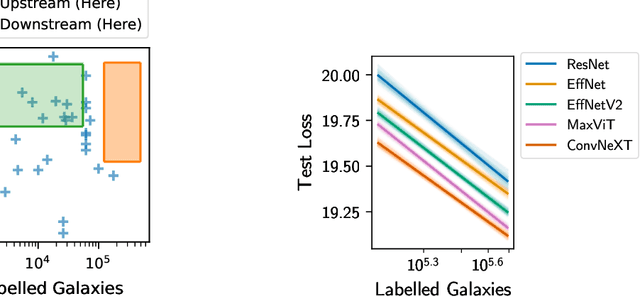
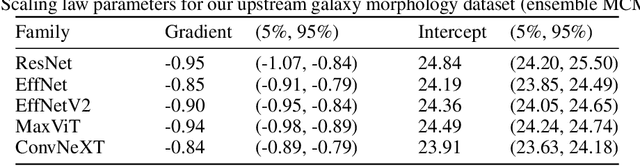
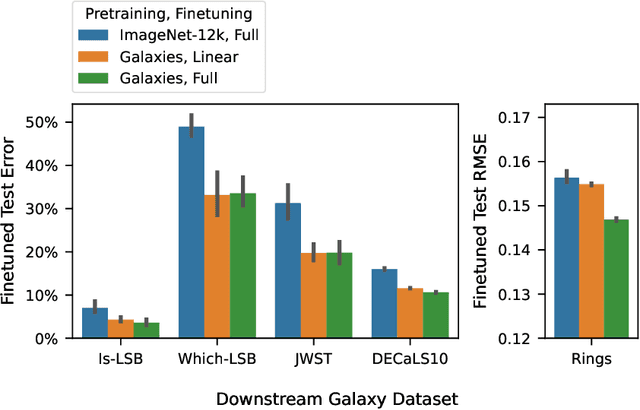
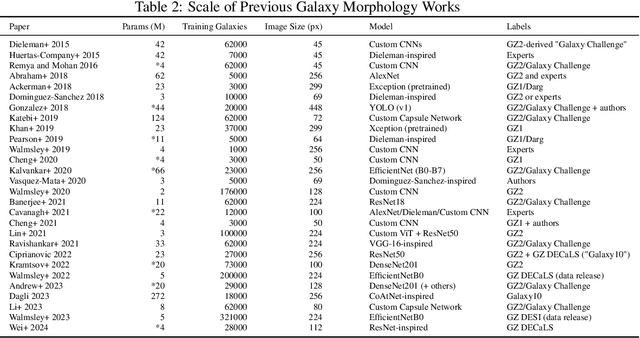
Abstract:We present the first systematic investigation of supervised scaling laws outside of an ImageNet-like context - on images of galaxies. We use 840k galaxy images and over 100M annotations by Galaxy Zoo volunteers, comparable in scale to Imagenet-1K. We find that adding annotated galaxy images provides a power law improvement in performance across all architectures and all tasks, while adding trainable parameters is effective only for some (typically more subjectively challenging) tasks. We then compare the downstream performance of finetuned models pretrained on either ImageNet-12k alone vs. additionally pretrained on our galaxy images. We achieve an average relative error rate reduction of 31% across 5 downstream tasks of scientific interest. Our finetuned models are more label-efficient and, unlike their ImageNet-12k-pretrained equivalents, often achieve linear transfer performance equal to that of end-to-end finetuning. We find relatively modest additional downstream benefits from scaling model size, implying that scaling alone is not sufficient to address our domain gap, and suggest that practitioners with qualitatively different images might benefit more from in-domain adaption followed by targeted downstream labelling.
Rapid prediction of lab-grown tissue properties using deep learning
Mar 31, 2023Abstract:The interactions between cells and the extracellular matrix are vital for the self-organisation of tissues. In this paper we present proof-of-concept to use machine learning tools to predict the role of this mechanobiology in the self-organisation of cell-laden hydrogels grown in tethered moulds. We develop a process for the automated generation of mould designs with and without key symmetries. We create a large training set with $N=6500$ cases by running detailed biophysical simulations of cell-matrix interactions using the contractile network dipole orientation (CONDOR) model for the self-organisation of cellular hydrogels within these moulds. These are used to train an implementation of the \texttt{pix2pix} deep learning model, reserving $740$ cases that were unseen in the training of the neural network for training and validation. Comparison between the predictions of the machine learning technique and the reserved predictions from the biophysical algorithm show that the machine learning algorithm makes excellent predictions. The machine learning algorithm is significantly faster than the biophysical method, opening the possibility of very high throughput rational design of moulds for pharmaceutical testing, regenerative medicine and fundamental studies of biology. Future extensions for scaffolds and 3D bioprinting will open additional applications.
Practical Galaxy Morphology Tools from Deep Supervised Representation Learning
Oct 25, 2021
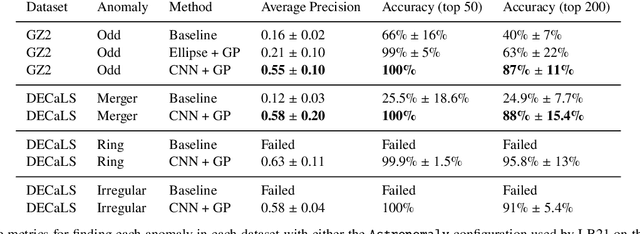
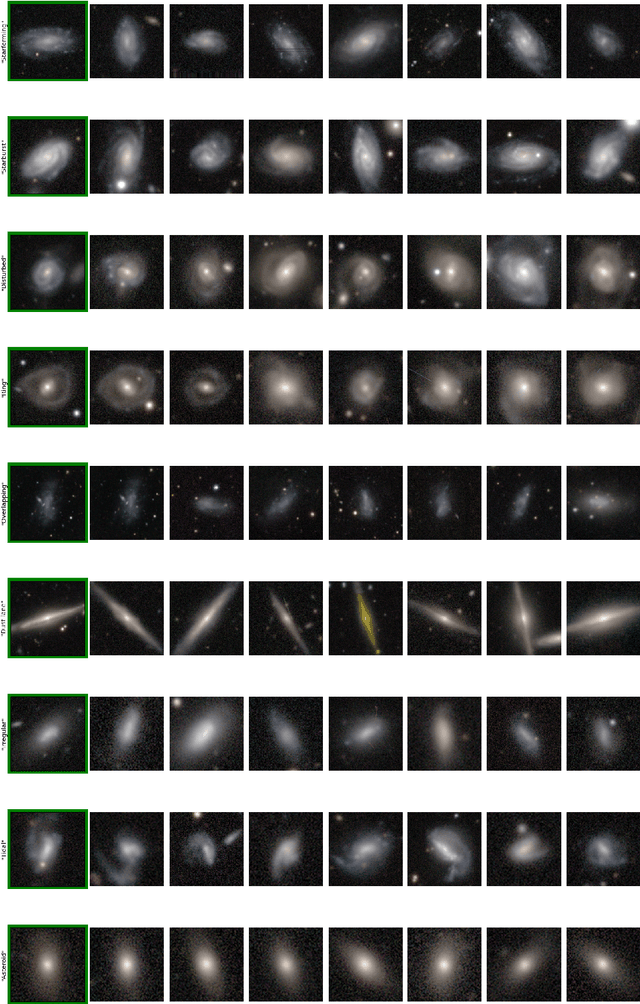
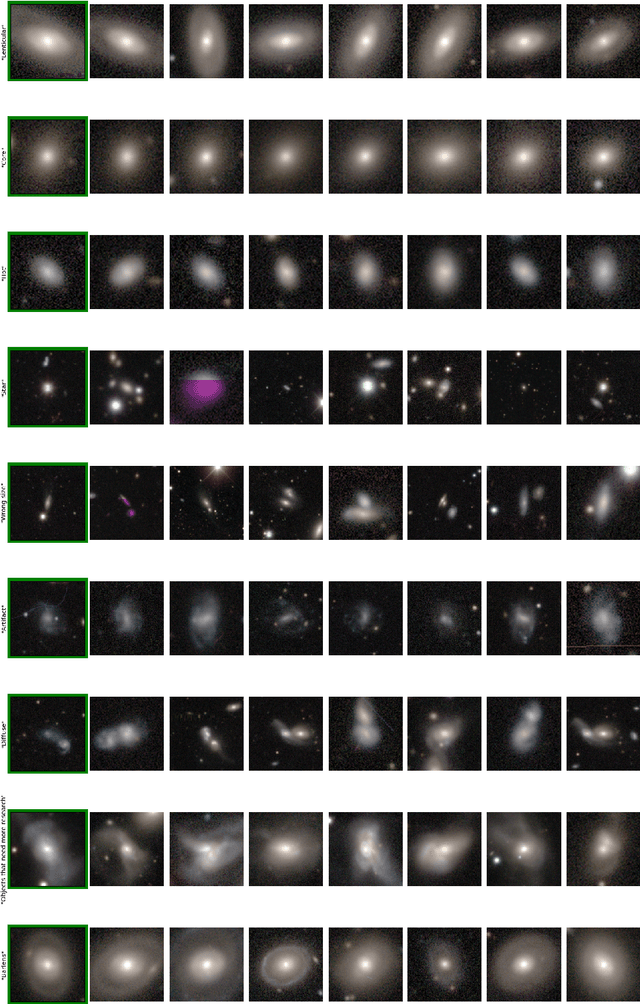
Abstract:Astronomers have typically set out to solve supervised machine learning problems by creating their own representations from scratch. We show that deep learning models trained to answer every Galaxy Zoo DECaLS question learn meaningful semantic representations of galaxies that are useful for new tasks on which the models were never trained. We exploit these representations to outperform existing approaches at several practical tasks crucial for investigating large galaxy samples. The first task is identifying galaxies of similar morphology to a query galaxy. Given a single galaxy assigned a free text tag by humans (e.g. `#diffuse'), we can find galaxies matching that tag for most tags. The second task is identifying the most interesting anomalies to a particular researcher. Our approach is 100\% accurate at identifying the most interesting 100 anomalies (as judged by Galaxy Zoo 2 volunteers). The third task is adapting a model to solve a new task using only a small number of newly-labelled galaxies. Models fine-tuned from our representation are better able to identify ring galaxies than models fine-tuned from terrestrial images (ImageNet) or trained from scratch. We solve each task with very few new labels; either one (for the similarity search) or several hundred (for anomaly detection or fine-tuning). This challenges the longstanding view that deep supervised methods require new large labelled datasets for practical use in astronomy. To help the community benefit from our pretrained models, we release our fine-tuning code zoobot. Zoobot is accessible to researchers with no prior experience in deep learning.
Galaxy Zoo: Probabilistic Morphology through Bayesian CNNs and Active Learning
May 17, 2019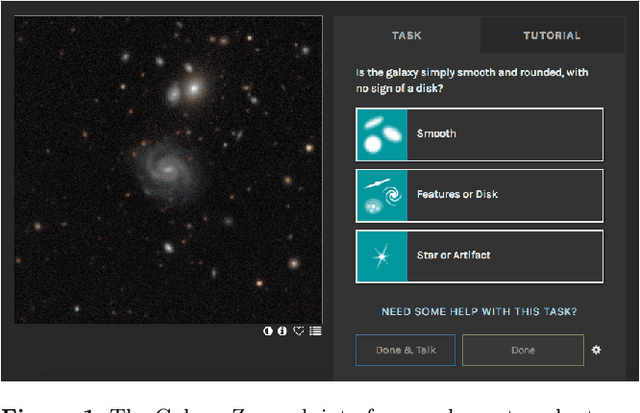

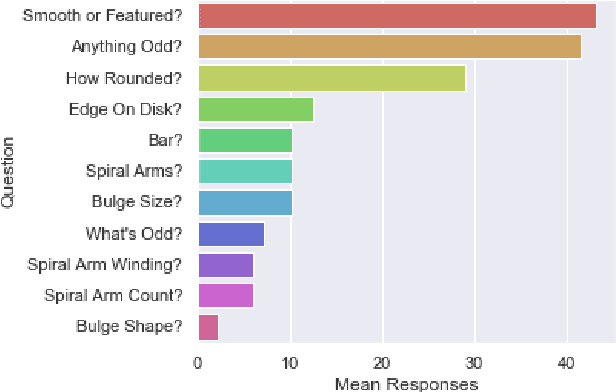

Abstract:We use Bayesian convolutional neural networks and a novel generative model of Galaxy Zoo volunteer responses to infer posteriors for the visual morphology of galaxies. Bayesian CNN can learn from galaxy images with uncertain labels and then, for previously unlabelled galaxies, predict the probability of each possible label. Our posteriors are well-calibrated (e.g. for predicting bars, we achieve coverage errors of 10.6% within 5 responses and 2.9% within 10 responses) and hence are reliable for practical use. Further, using our posteriors, we apply the active learning strategy BALD to request volunteer responses for the subset of galaxies which, if labelled, would be most informative for training our network. We show that training our Bayesian CNNs using active learning requires up to 35-60% fewer labelled galaxies, depending on the morphological feature being classified. By combining human and machine intelligence, Galaxy Zoo will be able to classify surveys of any conceivable scale on a timescale of weeks, providing massive and detailed morphology catalogues to support research into galaxy evolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge