Linwei Wang
Feasibility Study on Active Learning of Smart Surrogates for Scientific Simulations
Jul 10, 2024
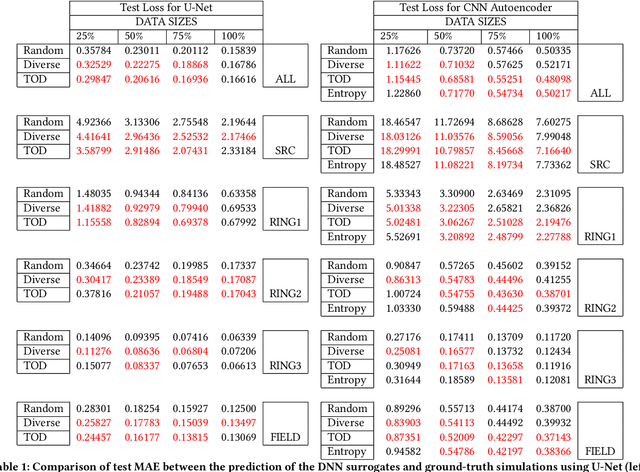


Abstract:High-performance scientific simulations, important for comprehension of complex systems, encounter computational challenges especially when exploring extensive parameter spaces. There has been an increasing interest in developing deep neural networks (DNNs) as surrogate models capable of accelerating the simulations. However, existing approaches for training these DNN surrogates rely on extensive simulation data which are heuristically selected and generated with expensive computation -- a challenge under-explored in the literature. In this paper, we investigate the potential of incorporating active learning into DNN surrogate training. This allows intelligent and objective selection of training simulations, reducing the need to generate extensive simulation data as well as the dependency of the performance of DNN surrogates on pre-defined training simulations. In the problem context of constructing DNN surrogates for diffusion equations with sources, we examine the efficacy of diversity- and uncertainty-based strategies for selecting training simulations, considering two different DNN architecture. The results set the groundwork for developing the high-performance computing infrastructure for Smart Surrogates that supports on-the-fly generation of simulation data steered by active learning strategies to potentially improve the efficiency of scientific simulations.
STLLaVA-Med: Self-Training Large Language and Vision Assistant for Medical
Jun 28, 2024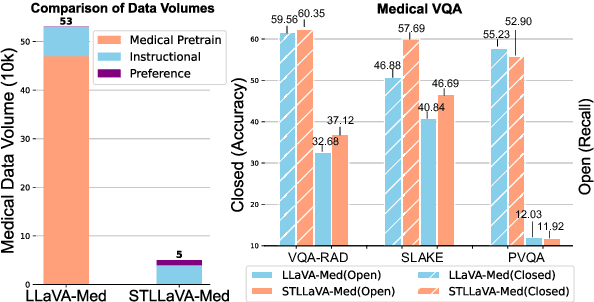

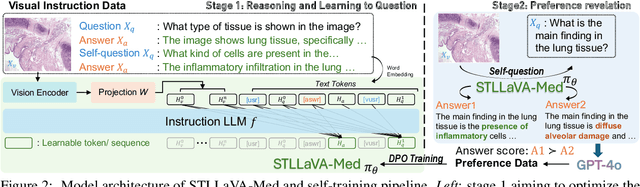
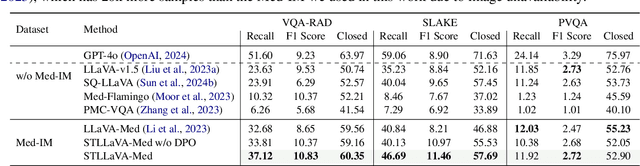
Abstract:Large Vision-Language Models (LVLMs) have shown significant potential in assisting medical diagnosis by leveraging extensive biomedical datasets. However, the advancement of medical image understanding and reasoning critically depends on building high-quality visual instruction data, which is costly and labor-intensive to obtain, particularly in the medical domain. To mitigate this data-starving issue, we introduce Self-Training Large Language and Vision Assistant for Medical (STLLaVA-Med). The proposed method is designed to train a policy model (an LVLM) capable of auto-generating medical visual instruction data to improve data efficiency, guided through Direct Preference Optimization (DPO). Specifically, a more powerful and larger LVLM (e.g., GPT-4o) is involved as a biomedical expert to oversee the DPO fine-tuning process on the auto-generated data, encouraging the policy model to align efficiently with human preferences. We validate the efficacy and data efficiency of STLLaVA-Med across three major medical Visual Question Answering (VQA) benchmarks, demonstrating competitive zero-shot performance with the utilization of only 9% of the medical data.
HyPer-EP: Meta-Learning Hybrid Personalized Models for Cardiac Electrophysiology
Mar 15, 2024


Abstract:Personalized virtual heart models have demonstrated increasing potential for clinical use, although the estimation of their parameters given patient-specific data remain a challenge. Traditional physics-based modeling approaches are computationally costly and often neglect the inherent structural errors in these models due to model simplifications and assumptions. Modern deep learning approaches, on the other hand, rely heavily on data supervision and lacks interpretability. In this paper, we present a novel hybrid modeling framework to describe a personalized cardiac digital twin as a combination of a physics-based known expression augmented by neural network modeling of its unknown gap to reality. We then present a novel meta-learning framework to enable the separate identification of both the physics-based and neural components in the hybrid model. We demonstrate the feasibility and generality of this hybrid modeling framework with two examples of instantiations and their proof-of-concept in synthetic experiments.
Unsupervised Learning of Hybrid Latent Dynamics: A Learn-to-Identify Framework
Mar 13, 2024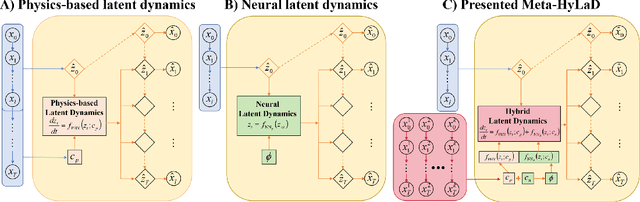
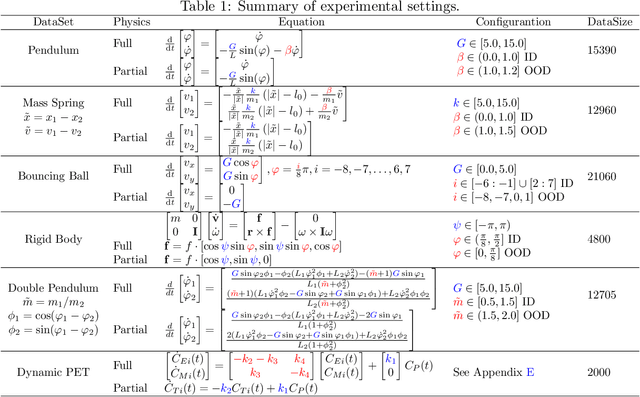
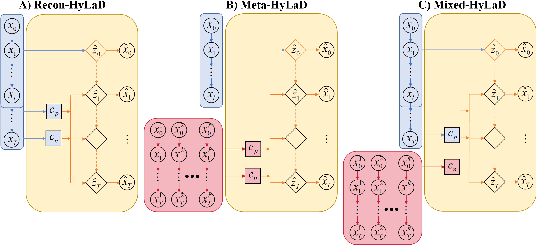
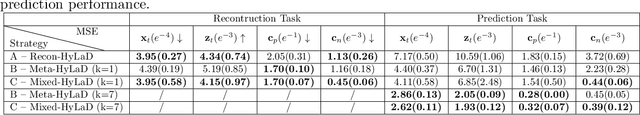
Abstract:Modern applications increasingly require unsupervised learning of latent dynamics from high-dimensional time-series. This presents a significant challenge of identifiability: many abstract latent representations may reconstruct observations, yet do they guarantee an adequate identification of the governing dynamics? This paper investigates this challenge from two angles: the use of physics inductive bias specific to the data being modeled, and a learn-to-identify strategy that separates forecasting objectives from the data used for the identification. We combine these two strategies in a novel framework for unsupervised meta-learning of hybrid latent dynamics (Meta-HyLaD) with: 1) a latent dynamic function that hybridize known mathematical expressions of prior physics with neural functions describing its unknown errors, and 2) a meta-learning formulation to learn to separately identify both components of the hybrid dynamics. Through extensive experiments on five physics and one biomedical systems, we provide strong evidence for the benefits of Meta-HyLaD to integrate rich prior knowledge while identifying their gap to observed data.
Hybrid Kinetics Embedding Framework for Dynamic PET Reconstruction
Mar 12, 2024Abstract:In dynamic positron emission tomography (PET) reconstruction, the importance of leveraging the temporal dependence of the data has been well appreciated. Current deep-learning solutions can be categorized in two groups in the way the temporal dynamics is modeled: data-driven approaches use spatiotemporal neural networks to learn the temporal dynamics of tracer kinetics from data, which relies heavily on data supervision; physics-based approaches leverage \textit{a priori} tracer kinetic models to focus on inferring their parameters, which relies heavily on the accuracy of the prior kinetic model. In this paper, we marry the strengths of these two approaches in a hybrid kinetics embedding (HyKE-Net) framework for dynamic PET reconstruction. We first introduce a novel \textit{hybrid} model of tracer kinetics consisting of a physics-based function augmented by a neural component to account for its gap to data-generating tracer kinetics, both identifiable from data. We then embed this hybrid model at the latent space of an encoding-decoding framework to enable both supervised and unsupervised identification of the hybrid kinetics and thereby dynamic PET reconstruction. Through both phantom and real-data experiments, we demonstrate the benefits of HyKE-Net -- especially in unsupervised reconstructions -- over existing physics-based and data-driven baselines as well as its ablated formulations where the embedded tracer kinetics are purely physics-based, purely neural, or hybrid but with a non-adaptable neural component.
LIBR+: Improving Intraoperative Liver Registration by Learning the Residual of Biomechanics-Based Deformable Registration
Mar 11, 2024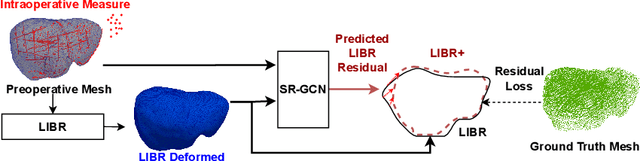

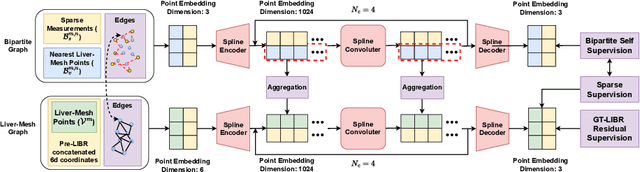

Abstract:The surgical environment imposes unique challenges to the intraoperative registration of organ shapes to their preoperatively-imaged geometry. Biomechanical model-based registration remains popular, while deep learning solutions remain limited due to the sparsity and variability of intraoperative measurements and the limited ground-truth deformation of an organ that can be obtained during the surgery. In this paper, we propose a novel \textit{hybrid} registration approach that leverage a linearized iterative boundary reconstruction (LIBR) method based on linear elastic biomechanics, and use deep neural networks to learn its residual to the ground-truth deformation (LIBR+). We further formulate a dual-branch spline-residual graph convolutional neural network (SR-GCN) to assimilate information from sparse and variable intraoperative measurements and effectively propagate it through the geometry of the 3D organ. Experiments on a large intraoperative liver registration dataset demonstrated the consistent improvements achieved by LIBR+ in comparison to existing rigid, biomechnical model-based non-rigid, and deep-learning based non-rigid approaches to intraoperative liver registration.
Distributionally Robust Optimization and Invariant Representation Learning for Addressing Subgroup Underrepresentation: Mechanisms and Limitations
Aug 12, 2023Abstract:Spurious correlation caused by subgroup underrepresentation has received increasing attention as a source of bias that can be perpetuated by deep neural networks (DNNs). Distributionally robust optimization has shown success in addressing this bias, although the underlying working mechanism mostly relies on upweighting under-performing samples as surrogates for those underrepresented in data. At the same time, while invariant representation learning has been a powerful choice for removing nuisance-sensitive features, it has been little considered in settings where spurious correlations are caused by significant underrepresentation of subgroups. In this paper, we take the first step to better understand and improve the mechanisms for debiasing spurious correlation due to subgroup underrepresentation in medical image classification. Through a comprehensive evaluation study, we first show that 1) generalized reweighting of under-performing samples can be problematic when bias is not the only cause for poor performance, while 2) naive invariant representation learning suffers from spurious correlations itself. We then present a novel approach that leverages robust optimization to facilitate the learning of invariant representations at the presence of spurious correlations. Finetuned classifiers utilizing such representation demonstrated improved abilities to reduce subgroup performance disparity, while maintaining high average and worst-group performance.
Learning Transferable Object-Centric Diffeomorphic Transformations for Data Augmentation in Medical Image Segmentation
Jul 25, 2023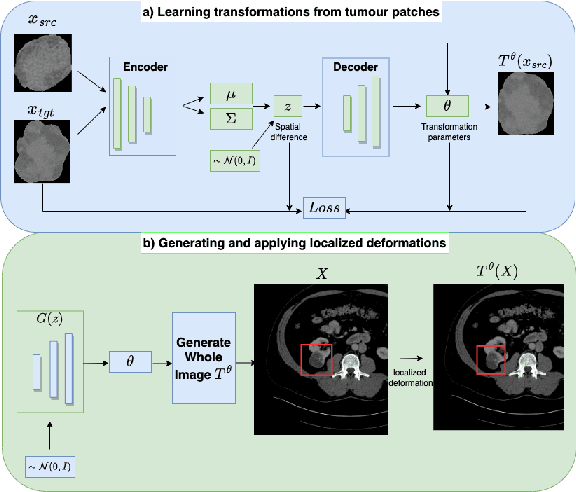
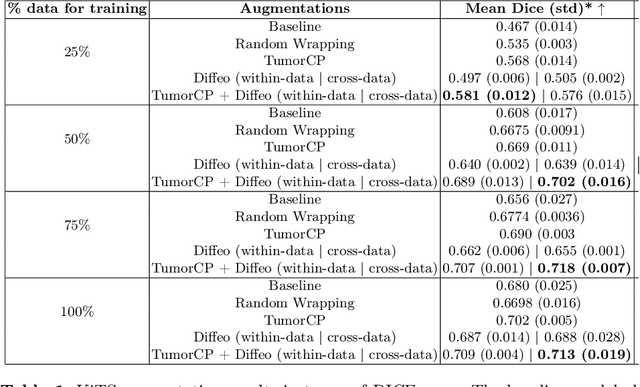
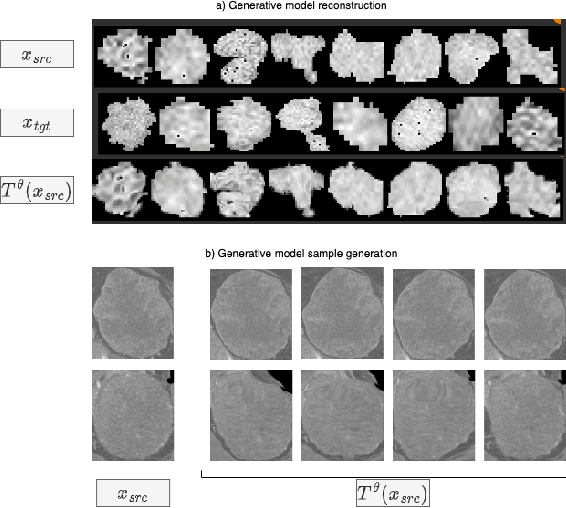
Abstract:Obtaining labelled data in medical image segmentation is challenging due to the need for pixel-level annotations by experts. Recent works have shown that augmenting the object of interest with deformable transformations can help mitigate this challenge. However, these transformations have been learned globally for the image, limiting their transferability across datasets or applicability in problems where image alignment is difficult. While object-centric augmentations provide a great opportunity to overcome these issues, existing works are only focused on position and random transformations without considering shape variations of the objects. To this end, we propose a novel object-centric data augmentation model that is able to learn the shape variations for the objects of interest and augment the object in place without modifying the rest of the image. We demonstrated its effectiveness in improving kidney tumour segmentation when leveraging shape variations learned both from within the same dataset and transferred from external datasets.
Interpretable Modeling and Reduction of Unknown Errors in Mechanistic Operators
Nov 02, 2022Abstract:Prior knowledge about the imaging physics provides a mechanistic forward operator that plays an important role in image reconstruction, although myriad sources of possible errors in the operator could negatively impact the reconstruction solutions. In this work, we propose to embed the traditional mechanistic forward operator inside a neural function, and focus on modeling and correcting its unknown errors in an interpretable manner. This is achieved by a conditional generative model that transforms a given mechanistic operator with unknown errors, arising from a latent space of self-organizing clusters of potential sources of error generation. Once learned, the generative model can be used in place of a fixed forward operator in any traditional optimization-based reconstruction process where, together with the inverse solution, the error in prior mechanistic forward operator can be minimized and the potential source of error uncovered. We apply the presented method to the reconstruction of heart electrical potential from body surface potential. In controlled simulation experiments and in-vivo real data experiments, we demonstrate that the presented method allowed reduction of errors in the physics-based forward operator and thereby delivered inverse reconstruction of heart-surface potential with increased accuracy.
* 11 pages, Conference: Medical Image Computing and Computer Assisted Intervention
Few-shot Generation of Personalized Neural Surrogates for Cardiac Simulation via Bayesian Meta-Learning
Oct 06, 2022Abstract:Clinical adoption of personalized virtual heart simulations faces challenges in model personalization and expensive computation. While an ideal solution is an efficient neural surrogate that at the same time is personalized to an individual subject, the state-of-the-art is either concerned with personalizing an expensive simulation model, or learning an efficient yet generic surrogate. This paper presents a completely new concept to achieve personalized neural surrogates in a single coherent framework of meta-learning (metaPNS). Instead of learning a single neural surrogate, we pursue the process of learning a personalized neural surrogate using a small amount of context data from a subject, in a novel formulation of few-shot generative modeling underpinned by: 1) a set-conditioned neural surrogate for cardiac simulation that, conditioned on subject-specific context data, learns to generate query simulations not included in the context set, and 2) a meta-model of amortized variational inference that learns to condition the neural surrogate via simple feed-forward embedding of context data. As test time, metaPNS delivers a personalized neural surrogate by fast feed-forward embedding of a small and flexible number of data available from an individual, achieving -- for the first time -- personalization and surrogate construction for expensive simulations in one end-to-end learning framework. Synthetic and real-data experiments demonstrated that metaPNS was able to improve personalization and predictive accuracy in comparison to conventionally-optimized cardiac simulation models, at a fraction of computation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge