Pradeep Bajracharya
Feasibility Study on Active Learning of Smart Surrogates for Scientific Simulations
Jul 10, 2024
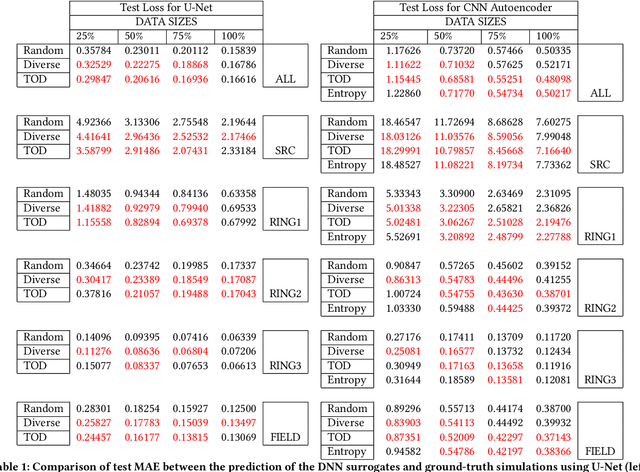


Abstract:High-performance scientific simulations, important for comprehension of complex systems, encounter computational challenges especially when exploring extensive parameter spaces. There has been an increasing interest in developing deep neural networks (DNNs) as surrogate models capable of accelerating the simulations. However, existing approaches for training these DNN surrogates rely on extensive simulation data which are heuristically selected and generated with expensive computation -- a challenge under-explored in the literature. In this paper, we investigate the potential of incorporating active learning into DNN surrogate training. This allows intelligent and objective selection of training simulations, reducing the need to generate extensive simulation data as well as the dependency of the performance of DNN surrogates on pre-defined training simulations. In the problem context of constructing DNN surrogates for diffusion equations with sources, we examine the efficacy of diversity- and uncertainty-based strategies for selecting training simulations, considering two different DNN architecture. The results set the groundwork for developing the high-performance computing infrastructure for Smart Surrogates that supports on-the-fly generation of simulation data steered by active learning strategies to potentially improve the efficiency of scientific simulations.
Fast Posterior Estimation of Cardiac Electrophysiological Model Parameters via Bayesian Active Learning
Oct 13, 2021



Abstract:Probabilistic estimation of cardiac electrophysiological model parameters serves an important step towards model personalization and uncertain quantification. The expensive computation associated with these model simulations, however, makes direct Markov Chain Monte Carlo (MCMC) sampling of the posterior probability density function (pdf) of model parameters computationally intensive. Approximated posterior pdfs resulting from replacing the simulation model with a computationally efficient surrogate, on the other hand, have seen limited accuracy. In this paper, we present a Bayesian active learning method to directly approximate the posterior pdf function of cardiac model parameters, in which we intelligently select training points to query the simulation model in order to learn the posterior pdf using a small number of samples. We integrate a generative model into Bayesian active learning to allow approximating posterior pdf of high-dimensional model parameters at the resolution of the cardiac mesh. We further introduce new acquisition functions to focus the selection of training points on better approximating the shape rather than the modes of the posterior pdf of interest. We evaluated the presented method in estimating tissue excitability in a 3D cardiac electrophysiological model in a range of synthetic and real-data experiments. We demonstrated its improved accuracy in approximating the posterior pdf compared to Bayesian active learning using regular acquisition functions, and substantially reduced computational cost in comparison to existing standard or accelerated MCMC sampling.
Semi-supervised Medical Image Classification with Global Latent Mixing
May 22, 2020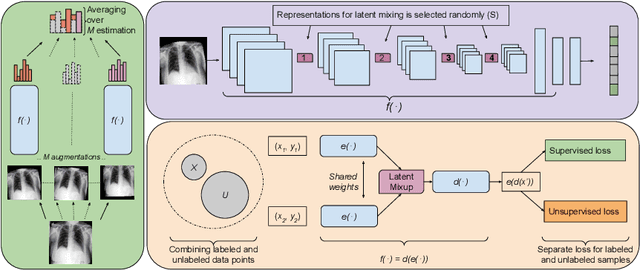
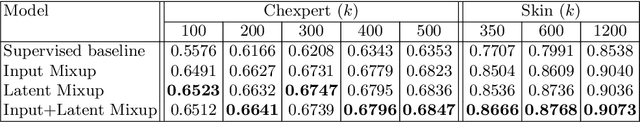
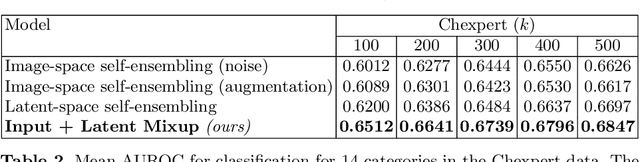
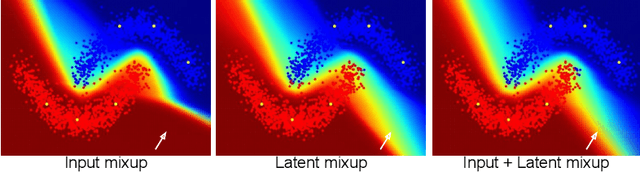
Abstract:Computer-aided diagnosis via deep learning relies on large-scale annotated data sets, which can be costly when involving expert knowledge. Semi-supervised learning (SSL) mitigates this challenge by leveraging unlabeled data. One effective SSL approach is to regularize the local smoothness of neural functions via perturbations around single data points. In this work, we argue that regularizing the global smoothness of neural functions by filling the void in between data points can further improve SSL. We present a novel SSL approach that trains the neural network on linear mixing of labeled and unlabeled data, at both the input and latent space in order to regularize different portions of the network. We evaluated the presented model on two distinct medical image data sets for semi-supervised classification of thoracic disease and skin lesion, demonstrating its improved performance over SSL with local perturbations and SSL with global mixing but at the input space only. Our code is available at https://github.com/Prasanna1991/LatentMixing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge