Natalia A. Trayanova
Fast Posterior Estimation of Cardiac Electrophysiological Model Parameters via Bayesian Active Learning
Oct 13, 2021



Abstract:Probabilistic estimation of cardiac electrophysiological model parameters serves an important step towards model personalization and uncertain quantification. The expensive computation associated with these model simulations, however, makes direct Markov Chain Monte Carlo (MCMC) sampling of the posterior probability density function (pdf) of model parameters computationally intensive. Approximated posterior pdfs resulting from replacing the simulation model with a computationally efficient surrogate, on the other hand, have seen limited accuracy. In this paper, we present a Bayesian active learning method to directly approximate the posterior pdf function of cardiac model parameters, in which we intelligently select training points to query the simulation model in order to learn the posterior pdf using a small number of samples. We integrate a generative model into Bayesian active learning to allow approximating posterior pdf of high-dimensional model parameters at the resolution of the cardiac mesh. We further introduce new acquisition functions to focus the selection of training points on better approximating the shape rather than the modes of the posterior pdf of interest. We evaluated the presented method in estimating tissue excitability in a 3D cardiac electrophysiological model in a range of synthetic and real-data experiments. We demonstrated its improved accuracy in approximating the posterior pdf compared to Bayesian active learning using regular acquisition functions, and substantially reduced computational cost in comparison to existing standard or accelerated MCMC sampling.
Anatomically-Informed Deep Learning on Contrast-Enhanced Cardiac MRI for Scar Segmentation and Clinical Feature Extraction
Oct 21, 2020
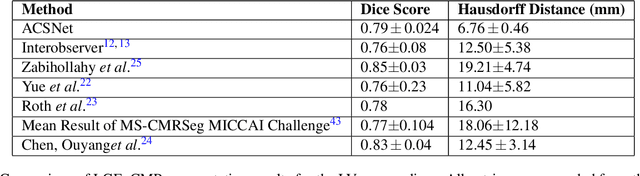


Abstract:Many cardiac diseases are associated with structural remodeling of the myocardium. Cardiac magnetic resonance (CMR) imaging with contrast enhancement, such as late gadolinium enhancement (LGE), has unparalleled capability to visualize fibrotic tissue remodeling, allowing for direct characterization of the pathophysiological abnormalities leading to arrhythmias and sudden cardiac death (SCD). Automating segmentation of the ventricles with fibrosis distribution could dramatically enhance the utility of LGE-CMR in heart disease clinical research and in the management of patients with risk of arrhythmias and SCD. Here we describe an anatomically-informed deep learning (DL) approach to myocardium and scar segmentation and clinical feature extraction from LGE-CMR images. The technology enables clinical use by ensuring anatomical accuracy and complete automation. Algorithm performance is strong for both myocardium segmentation ($98\%$ accuracy and $0.79$ Dice score in a hold-out test set) and evaluation measures shown to correlate with heart disease, such as scar amount ($6.3\%$ relative error). Our approach for clinical feature extraction, which satisfies highly complex geometric constraints without stunting the learning process, has the potential of a broad applicability in computer vision beyond cardiology, and even outside of medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge