John L. Sapp
Few-shot Generation of Personalized Neural Surrogates for Cardiac Simulation via Bayesian Meta-Learning
Oct 06, 2022Abstract:Clinical adoption of personalized virtual heart simulations faces challenges in model personalization and expensive computation. While an ideal solution is an efficient neural surrogate that at the same time is personalized to an individual subject, the state-of-the-art is either concerned with personalizing an expensive simulation model, or learning an efficient yet generic surrogate. This paper presents a completely new concept to achieve personalized neural surrogates in a single coherent framework of meta-learning (metaPNS). Instead of learning a single neural surrogate, we pursue the process of learning a personalized neural surrogate using a small amount of context data from a subject, in a novel formulation of few-shot generative modeling underpinned by: 1) a set-conditioned neural surrogate for cardiac simulation that, conditioned on subject-specific context data, learns to generate query simulations not included in the context set, and 2) a meta-model of amortized variational inference that learns to condition the neural surrogate via simple feed-forward embedding of context data. As test time, metaPNS delivers a personalized neural surrogate by fast feed-forward embedding of a small and flexible number of data available from an individual, achieving -- for the first time -- personalization and surrogate construction for expensive simulations in one end-to-end learning framework. Synthetic and real-data experiments demonstrated that metaPNS was able to improve personalization and predictive accuracy in comparison to conventionally-optimized cardiac simulation models, at a fraction of computation.
Fast Posterior Estimation of Cardiac Electrophysiological Model Parameters via Bayesian Active Learning
Oct 13, 2021



Abstract:Probabilistic estimation of cardiac electrophysiological model parameters serves an important step towards model personalization and uncertain quantification. The expensive computation associated with these model simulations, however, makes direct Markov Chain Monte Carlo (MCMC) sampling of the posterior probability density function (pdf) of model parameters computationally intensive. Approximated posterior pdfs resulting from replacing the simulation model with a computationally efficient surrogate, on the other hand, have seen limited accuracy. In this paper, we present a Bayesian active learning method to directly approximate the posterior pdf function of cardiac model parameters, in which we intelligently select training points to query the simulation model in order to learn the posterior pdf using a small number of samples. We integrate a generative model into Bayesian active learning to allow approximating posterior pdf of high-dimensional model parameters at the resolution of the cardiac mesh. We further introduce new acquisition functions to focus the selection of training points on better approximating the shape rather than the modes of the posterior pdf of interest. We evaluated the presented method in estimating tissue excitability in a 3D cardiac electrophysiological model in a range of synthetic and real-data experiments. We demonstrated its improved accuracy in approximating the posterior pdf compared to Bayesian active learning using regular acquisition functions, and substantially reduced computational cost in comparison to existing standard or accelerated MCMC sampling.
Quantifying the Uncertainty in Model Parameters Using Gaussian Process-Based Markov Chain Monte Carlo: An Application to Cardiac Electrophysiological Models
Jun 02, 2020



Abstract:Estimation of patient-specific model parameters is important for personalized modeling, although sparse and noisy clinical data can introduce significant uncertainty in the estimated parameter values. This importance source of uncertainty, if left unquantified, will lead to unknown variability in model outputs that hinder their reliable adoptions. Probabilistic estimation model parameters, however, remains an unresolved challenge because standard Markov Chain Monte Carlo sampling requires repeated model simulations that are computationally infeasible. A common solution is to replace the simulation model with a computationally-efficient surrogate for a faster sampling. However, by sampling from an approximation of the exact posterior probability density function (pdf) of the parameters, the efficiency is gained at the expense of sampling accuracy. In this paper, we address this issue by integrating surrogate modeling into Metropolis Hasting (MH) sampling of the exact posterior pdfs to improve its acceptance rate. It is done by first quickly constructing a Gaussian process (GP) surrogate of the exact posterior pdfs using deterministic optimization. This efficient surrogate is then used to modify commonly-used proposal distributions in MH sampling such that only proposals accepted by the surrogate will be tested by the exact posterior pdf for acceptance/rejection, reducing unnecessary model simulations at unlikely candidates. Synthetic and real-data experiments using the presented method show a significant gain in computational efficiency without compromising the accuracy. In addition, insights into the non-identifiability and heterogeneity of tissue properties can be gained from the obtained posterior distributions.
High-dimensional Bayesian Optimization of Personalized Cardiac Model Parameters via an Embedded Generative Model
May 15, 2020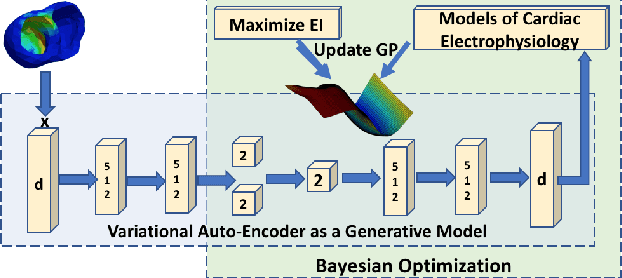
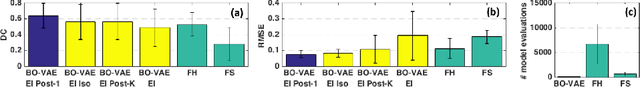
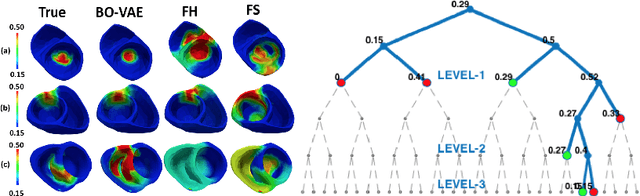
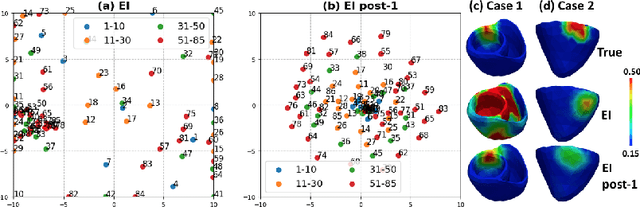
Abstract:The estimation of patient-specific tissue properties in the form of model parameters is important for personalized physiological models. However, these tissue properties are spatially varying across the underlying anatomical model, presenting a significance challenge of high-dimensional (HD) optimization at the presence of limited measurement data. A common solution to reduce the dimension of the parameter space is to explicitly partition the anatomical mesh, either into a fixed small number of segments or a multi-scale hierarchy. This anatomy-based reduction of parameter space presents a fundamental bottleneck to parameter estimation, resulting in solutions that are either too low in resolution to reflect tissue heterogeneity, or too high in dimension to be reliably estimated within feasible computation. In this paper, we present a novel concept that embeds a generative variational auto-encoder (VAE) into the objective function of Bayesian optimization, providing an implicit low-dimensional (LD) search space that represents the generative code of the HD spatially-varying tissue properties. In addition, the VAE-encoded knowledge about the generative code is further used to guide the exploration of the search space. The presented method is applied to estimating tissue excitability in a cardiac electrophysiological model. Synthetic and real-data experiments demonstrate its ability to improve the accuracy of parameter estimation with more than 10x gain in efficiency.
Bayesian Optimization on Large Graphs via a Graph Convolutional Generative Model: Application in Cardiac Model Personalization
Jul 01, 2019



Abstract:Personalization of cardiac models involves the optimization of organ tissue properties that vary spatially over the non-Euclidean geometry model of the heart. To represent the high-dimensional (HD) unknown of tissue properties, most existing works rely on a low-dimensional (LD) partitioning of the geometrical model. While this exploits the geometry of the heart, it is of limited expressiveness to allow partitioning that is small enough for effective optimization. Recently, a variational auto-encoder (VAE) was utilized as a more expressive generative model to embed the HD optimization into the LD latent space. Its Euclidean nature, however, neglects the rich geometrical information in the heart. In this paper, we present a novel graph convolutional VAE to allow generative modeling of non-Euclidean data, and utilize it to embed Bayesian optimization of large graphs into a small latent space. This approach bridges the gap of previous works by introducing an expressive generative model that is able to incorporate the knowledge of spatial proximity and hierarchical compositionality of the underlying geometry. It further allows transferring of the learned features across different geometries, which was not possible with a regular VAE. We demonstrate these benefits of the presented method in synthetic and real data experiments of estimating tissue excitability in a cardiac electrophysiological model.
Deep Generative Model with Beta Bernoulli Process for Modeling and Learning Confounding Factors
Oct 31, 2018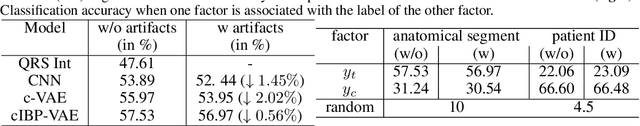
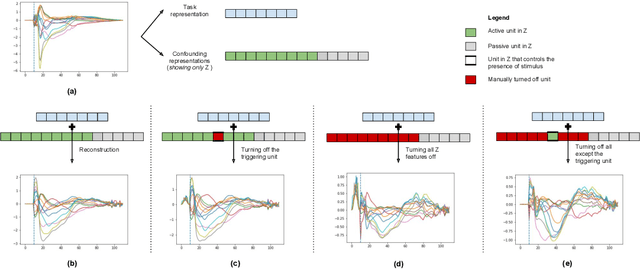
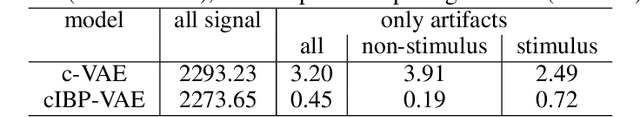
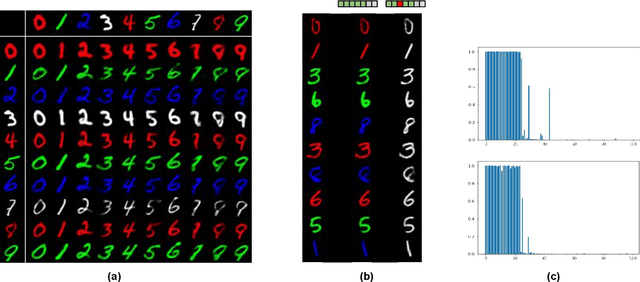
Abstract:While deep representation learning has become increasingly capable of separating task-relevant representations from other confounding factors in the data, two significant challenges remain. First, there is often an unknown and potentially infinite number of confounding factors coinciding in the data. Second, not all of these factors are readily observable. In this paper, we present a deep conditional generative model that learns to disentangle a task-relevant representation from an unknown number of confounding factors that may grow infinitely. This is achieved by marrying the representational power of deep generative models with Bayesian non-parametric factor models, where a supervised deterministic encoder learns task-related representation and a probabilistic encoder with an Indian Buffet Process (IBP) learns the unknown number of unobservable confounding factors. We tested the presented model in two datasets: a handwritten digit dataset (MNIST) augmented with colored digits and a clinical ECG dataset with significant inter-subject variations and augmented with signal artifacts. These diverse data sets highlighted the ability of the presented model to grow with the complexity of the data and identify the absence or presence of unobserved confounding factors.
Learning disentangled representation from 12-lead electrograms: application in localizing the origin of Ventricular Tachycardia
Aug 04, 2018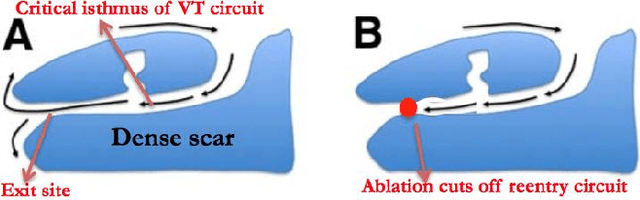
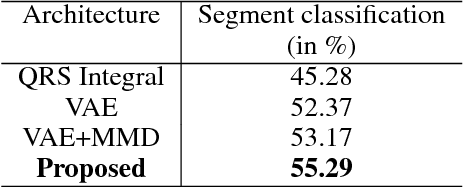
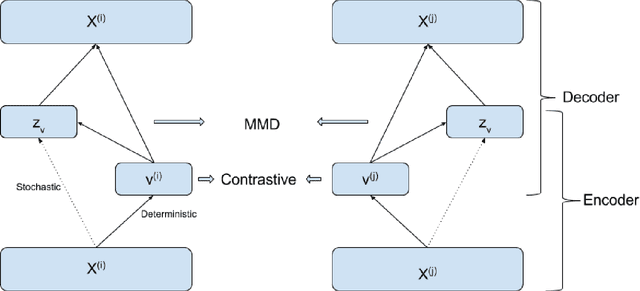
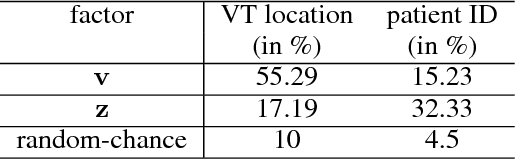
Abstract:The increasing availability of electrocardiogram (ECG) data has motivated the use of data-driven models for automating various clinical tasks based on ECG data. The development of subject-specific models are limited by the cost and difficulty of obtaining sufficient training data for each individual. The alternative of population model, however, faces challenges caused by the significant inter-subject variations within the ECG data. We address this challenge by investigating for the first time the problem of learning representations for clinically-informative variables while disentangling other factors of variations within the ECG data. In this work, we present a conditional variational autoencoder (VAE) to extract the subject-specific adjustment to the ECG data, conditioned on task-specific representations learned from a deterministic encoder. To encourage the representation for inter-subject variations to be independent from the task-specific representation, maximum mean discrepancy is used to match all the moments between the distributions learned by the VAE conditioning on the code from the deterministic encoder. The learning of the task-specific representation is regularized by a weak supervision in the form of contrastive regularization. We apply the proposed method to a novel yet important clinical task of classifying the origin of ventricular tachycardia (VT) into pre-defined segments, demonstrating the efficacy of the proposed method against the standard VAE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge