Learning disentangled representation from 12-lead electrograms: application in localizing the origin of Ventricular Tachycardia
Paper and Code
Aug 04, 2018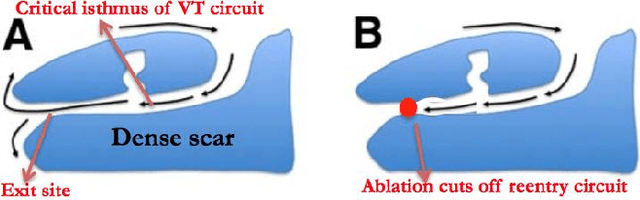
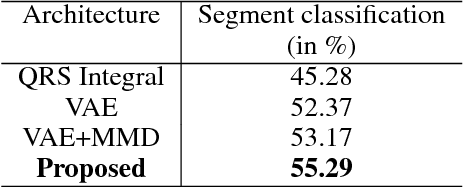
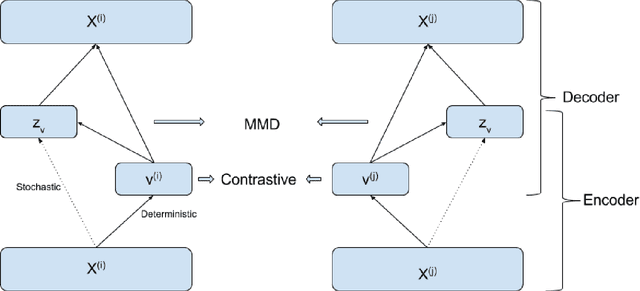
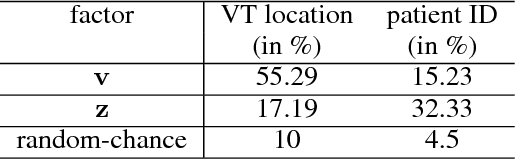
The increasing availability of electrocardiogram (ECG) data has motivated the use of data-driven models for automating various clinical tasks based on ECG data. The development of subject-specific models are limited by the cost and difficulty of obtaining sufficient training data for each individual. The alternative of population model, however, faces challenges caused by the significant inter-subject variations within the ECG data. We address this challenge by investigating for the first time the problem of learning representations for clinically-informative variables while disentangling other factors of variations within the ECG data. In this work, we present a conditional variational autoencoder (VAE) to extract the subject-specific adjustment to the ECG data, conditioned on task-specific representations learned from a deterministic encoder. To encourage the representation for inter-subject variations to be independent from the task-specific representation, maximum mean discrepancy is used to match all the moments between the distributions learned by the VAE conditioning on the code from the deterministic encoder. The learning of the task-specific representation is regularized by a weak supervision in the form of contrastive regularization. We apply the proposed method to a novel yet important clinical task of classifying the origin of ventricular tachycardia (VT) into pre-defined segments, demonstrating the efficacy of the proposed method against the standard VAE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge