Prashnna K Gyawali
Med-MMFL: A Multimodal Federated Learning Benchmark in Healthcare
Feb 04, 2026Abstract:Federated learning (FL) enables collaborative model training across decentralized medical institutions while preserving data privacy. However, medical FL benchmarks remain scarce, with existing efforts focusing mainly on unimodal or bimodal modalities and a limited range of medical tasks. This gap underscores the need for standardized evaluation to advance systematic understanding in medical MultiModal FL (MMFL). To this end, we introduce Med-MMFL, the first comprehensive MMFL benchmark for the medical domain, encompassing diverse modalities, tasks, and federation scenarios. Our benchmark evaluates six representative state-of-the-art FL algorithms, covering different aggregation strategies, loss formulations, and regularization techniques. It spans datasets with 2 to 4 modalities, comprising a total of 10 unique medical modalities, including text, pathology images, ECG, X-ray, radiology reports, and multiple MRI sequences. Experiments are conducted across naturally federated, synthetic IID, and synthetic non-IID settings to simulate real-world heterogeneity. We assess segmentation, classification, modality alignment (retrieval), and VQA tasks. To support reproducibility and fair comparison of future multimodal federated learning (MMFL) methods under realistic medical settings, we release the complete benchmark implementation, including data processing and partitioning pipelines, at https://github.com/bhattarailab/Med-MMFL-Benchmark .
Ensembling improves stability and power of feature selection for deep learning models
Oct 02, 2022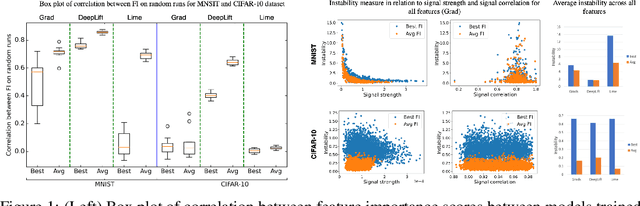
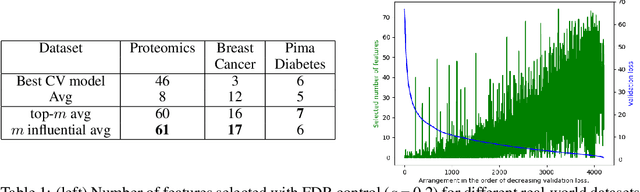
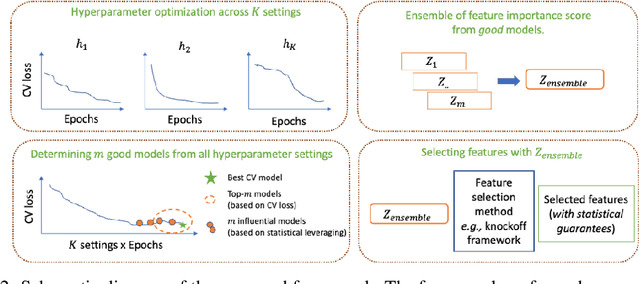
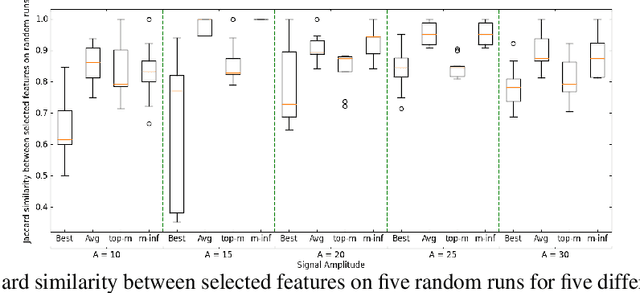
Abstract:With the growing adoption of deep learning models in different real-world domains, including computational biology, it is often necessary to understand which data features are essential for the model's decision. Despite extensive recent efforts to define different feature importance metrics for deep learning models, we identified that inherent stochasticity in the design and training of deep learning models makes commonly used feature importance scores unstable. This results in varied explanations or selections of different features across different runs of the model. We demonstrate how the signal strength of features and correlation among features directly contribute to this instability. To address this instability, we explore the ensembling of feature importance scores of models across different epochs and find that this simple approach can substantially address this issue. For example, we consider knockoff inference as they allow feature selection with statistical guarantees. We discover considerable variability in selected features in different epochs of deep learning training, and the best selection of features doesn't necessarily occur at the lowest validation loss, the conventional approach to determine the best model. As such, we present a framework to combine the feature importance of trained models across different hyperparameter settings and epochs, and instead of selecting features from one best model, we perform an ensemble of feature importance scores from numerous good models. Across the range of experiments in simulated and various real-world datasets, we demonstrate that the proposed framework consistently improves the power of feature selection.
Improving genetic risk prediction across diverse population by disentangling ancestry representations
May 10, 2022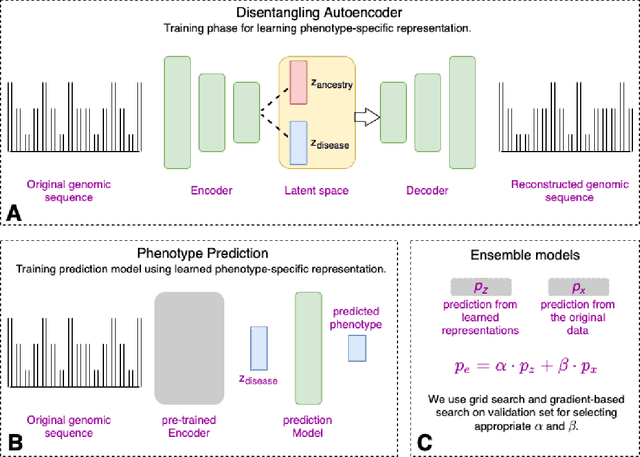
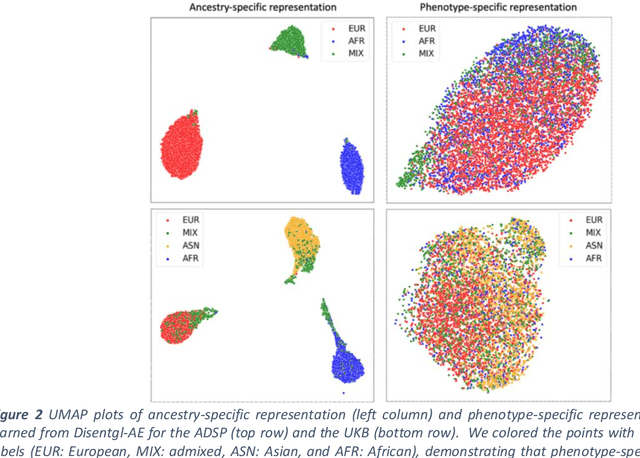
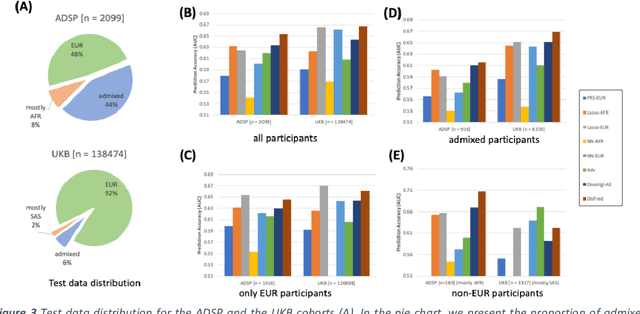
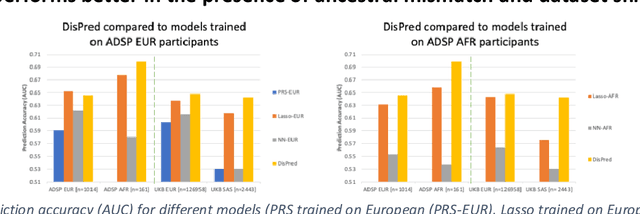
Abstract:Risk prediction models using genetic data have seen increasing traction in genomics. However, most of the polygenic risk models were developed using data from participants with similar (mostly European) ancestry. This can lead to biases in the risk predictors resulting in poor generalization when applied to minority populations and admixed individuals such as African Americans. To address this bias, largely due to the prediction models being confounded by the underlying population structure, we propose a novel deep-learning framework that leverages data from diverse population and disentangles ancestry from the phenotype-relevant information in its representation. The ancestry disentangled representation can be used to build risk predictors that perform better across minority populations. We applied the proposed method to the analysis of Alzheimer's disease genetics. Comparing with standard linear and nonlinear risk prediction methods, the proposed method substantially improves risk prediction in minority populations, particularly for admixed individuals.
Reliable Estimation of Kullback-Leibler Divergence by Controlling Discriminator Complexity in the Reproducing Kernel Hilbert Space
Mar 20, 2020
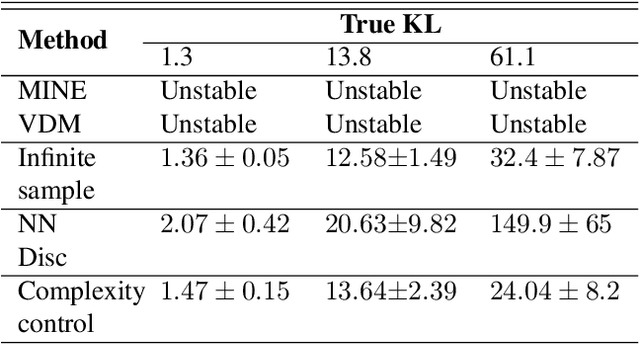
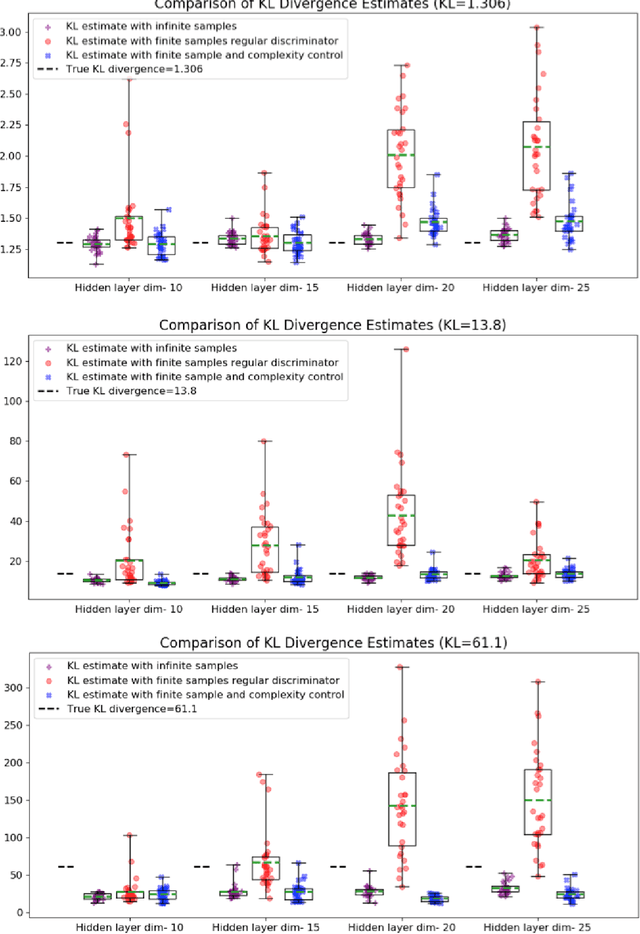
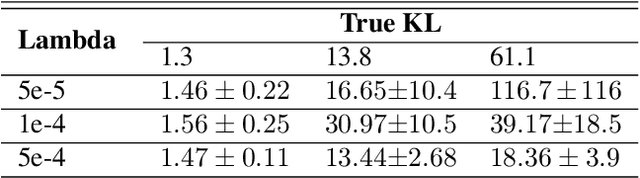
Abstract:Several scalable methods to compute the Kullback Leibler (KL) divergence between two distributions using their samples have been proposed and applied in large-scale machine learning models. While they have been found to be unstable, the theoretical root cause of the problem is not clear. In this paper, we study in detail a generative adversarial network based approach that uses a neural network discriminator to estimate KL divergence. We argue that, in such case, high fluctuations in the estimates are a consequence of not controlling the complexity of the discriminator function space. We provide a theoretical underpinning and remedy for this problem through the following contributions. First, we construct a discriminator in the Reproducing Kernel Hilbert Space (RKHS). This enables us to leverage sample complexity and mean embedding to theoretically relate the error probability bound of the KL estimates to the complexity of the neural-net discriminator. Based on this theory, we then present a scalable way to control the complexity of the discriminator for a consistent estimation of KL divergence. We support both our proposed theory and method to control the complexity of the RKHS discriminator in controlled experiments.
Wavelets to the Rescue: Improving Sample Quality of Latent Variable Deep Generative Models
Oct 26, 2019
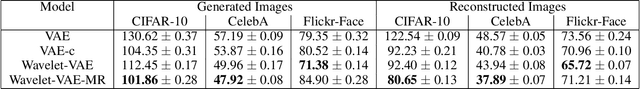
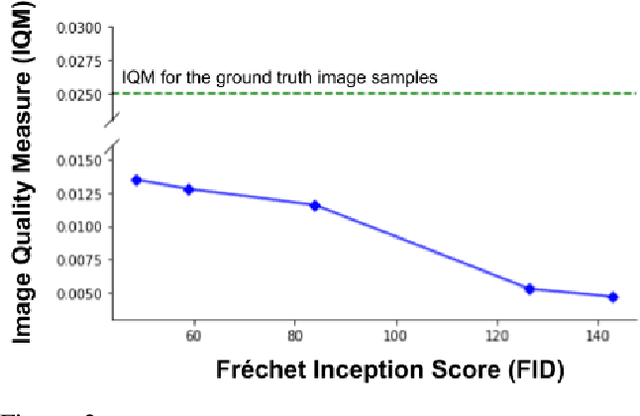
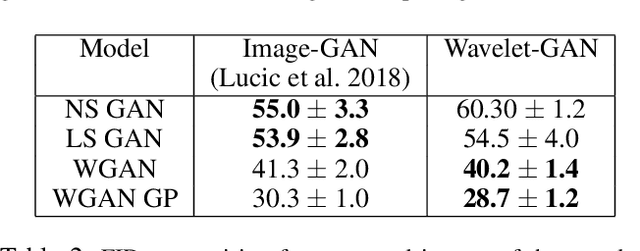
Abstract:Variational Autoencoders (VAE) are probabilistic deep generative models underpinned by elegant theory, stable training processes, and meaningful manifold representations. However, they produce blurry images due to a lack of explicit emphasis over high-frequency textural details of the images, and the difficulty to directly model the complex joint probability distribution over the high-dimensional image space. In this work, we approach these two challenges with a novel wavelet space VAE that uses the decoder to model the images in the wavelet coefficient space. This enables the VAE to emphasize over high-frequency components within an image obtained via wavelet decomposition. Additionally, by decomposing the complex function of generating high-dimensional images into inverse wavelet transformation and generation of wavelet coefficients, the latter becomes simpler to model by the VAE. We empirically validate that deep generative models operating in the wavelet space can generate images of higher quality than the image (RGB) space counterparts. Quantitatively, on benchmark natural image datasets, we achieve consistently better FID scores than VAE based architectures and competitive FID scores with a variety of GAN models for the same architectural and experimental setup. Furthermore, the proposed wavelet-based generative model retains desirable attributes like disentangled and informative latent representation without losing the quality in the generated samples.
Deep Generative Model with Beta Bernoulli Process for Modeling and Learning Confounding Factors
Oct 31, 2018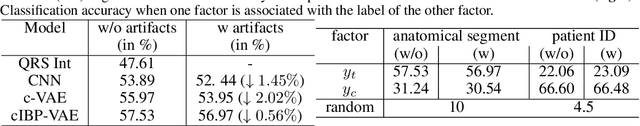
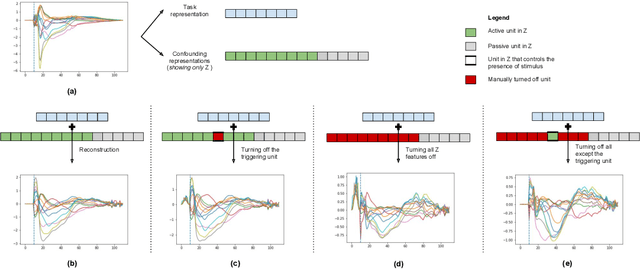
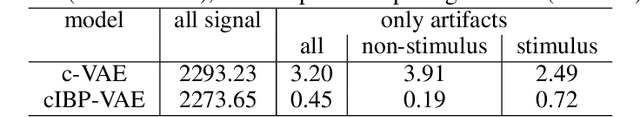
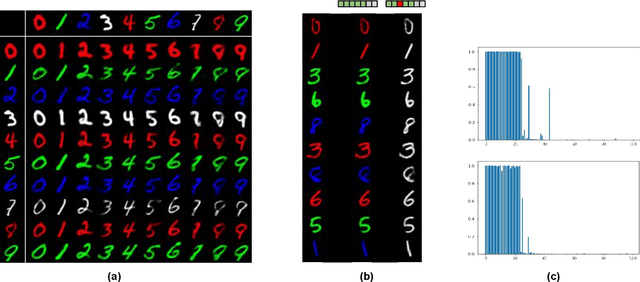
Abstract:While deep representation learning has become increasingly capable of separating task-relevant representations from other confounding factors in the data, two significant challenges remain. First, there is often an unknown and potentially infinite number of confounding factors coinciding in the data. Second, not all of these factors are readily observable. In this paper, we present a deep conditional generative model that learns to disentangle a task-relevant representation from an unknown number of confounding factors that may grow infinitely. This is achieved by marrying the representational power of deep generative models with Bayesian non-parametric factor models, where a supervised deterministic encoder learns task-related representation and a probabilistic encoder with an Indian Buffet Process (IBP) learns the unknown number of unobservable confounding factors. We tested the presented model in two datasets: a handwritten digit dataset (MNIST) augmented with colored digits and a clinical ECG dataset with significant inter-subject variations and augmented with signal artifacts. These diverse data sets highlighted the ability of the presented model to grow with the complexity of the data and identify the absence or presence of unobserved confounding factors.
Learning disentangled representation from 12-lead electrograms: application in localizing the origin of Ventricular Tachycardia
Aug 04, 2018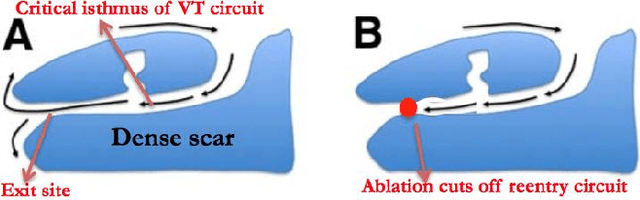
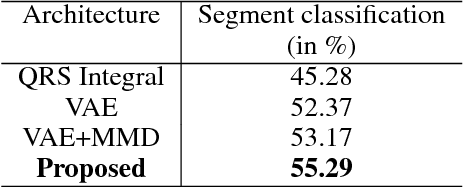
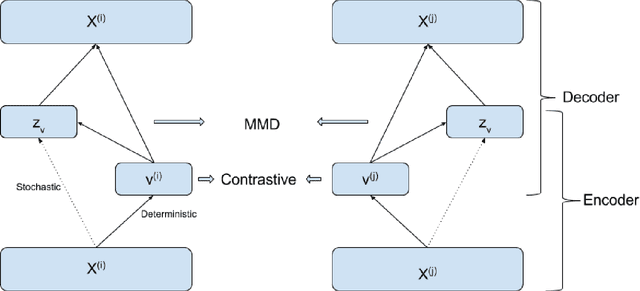
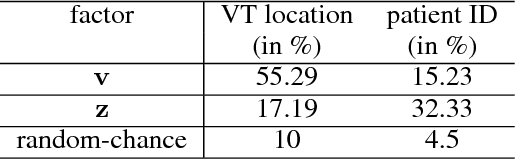
Abstract:The increasing availability of electrocardiogram (ECG) data has motivated the use of data-driven models for automating various clinical tasks based on ECG data. The development of subject-specific models are limited by the cost and difficulty of obtaining sufficient training data for each individual. The alternative of population model, however, faces challenges caused by the significant inter-subject variations within the ECG data. We address this challenge by investigating for the first time the problem of learning representations for clinically-informative variables while disentangling other factors of variations within the ECG data. In this work, we present a conditional variational autoencoder (VAE) to extract the subject-specific adjustment to the ECG data, conditioned on task-specific representations learned from a deterministic encoder. To encourage the representation for inter-subject variations to be independent from the task-specific representation, maximum mean discrepancy is used to match all the moments between the distributions learned by the VAE conditioning on the code from the deterministic encoder. The learning of the task-specific representation is regularized by a weak supervision in the form of contrastive regularization. We apply the proposed method to a novel yet important clinical task of classifying the origin of ventricular tachycardia (VT) into pre-defined segments, demonstrating the efficacy of the proposed method against the standard VAE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge