Ryan Missel
HyPer-EP: Meta-Learning Hybrid Personalized Models for Cardiac Electrophysiology
Mar 15, 2024


Abstract:Personalized virtual heart models have demonstrated increasing potential for clinical use, although the estimation of their parameters given patient-specific data remain a challenge. Traditional physics-based modeling approaches are computationally costly and often neglect the inherent structural errors in these models due to model simplifications and assumptions. Modern deep learning approaches, on the other hand, rely heavily on data supervision and lacks interpretability. In this paper, we present a novel hybrid modeling framework to describe a personalized cardiac digital twin as a combination of a physics-based known expression augmented by neural network modeling of its unknown gap to reality. We then present a novel meta-learning framework to enable the separate identification of both the physics-based and neural components in the hybrid model. We demonstrate the feasibility and generality of this hybrid modeling framework with two examples of instantiations and their proof-of-concept in synthetic experiments.
Unsupervised Learning of Hybrid Latent Dynamics: A Learn-to-Identify Framework
Mar 13, 2024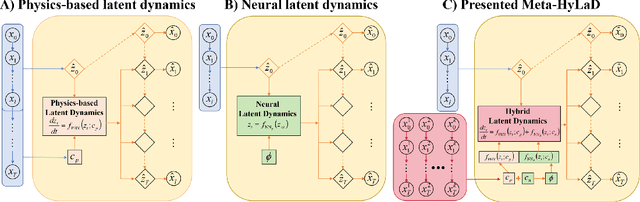
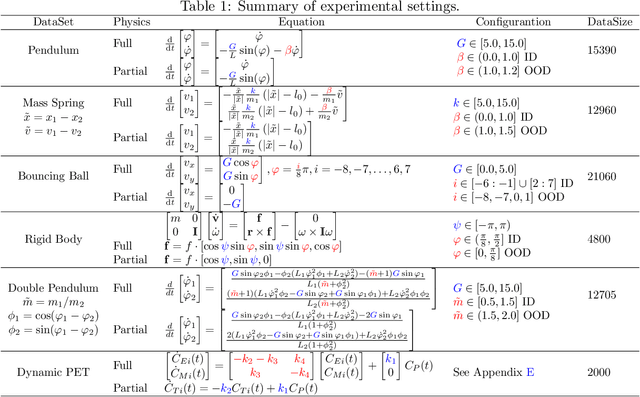
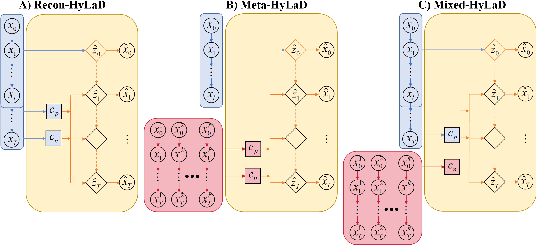
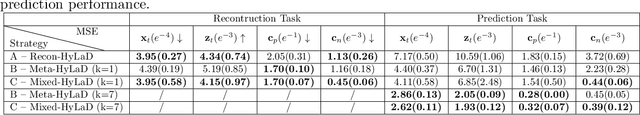
Abstract:Modern applications increasingly require unsupervised learning of latent dynamics from high-dimensional time-series. This presents a significant challenge of identifiability: many abstract latent representations may reconstruct observations, yet do they guarantee an adequate identification of the governing dynamics? This paper investigates this challenge from two angles: the use of physics inductive bias specific to the data being modeled, and a learn-to-identify strategy that separates forecasting objectives from the data used for the identification. We combine these two strategies in a novel framework for unsupervised meta-learning of hybrid latent dynamics (Meta-HyLaD) with: 1) a latent dynamic function that hybridize known mathematical expressions of prior physics with neural functions describing its unknown errors, and 2) a meta-learning formulation to learn to separately identify both components of the hybrid dynamics. Through extensive experiments on five physics and one biomedical systems, we provide strong evidence for the benefits of Meta-HyLaD to integrate rich prior knowledge while identifying their gap to observed data.
Few-shot Generation of Personalized Neural Surrogates for Cardiac Simulation via Bayesian Meta-Learning
Oct 06, 2022Abstract:Clinical adoption of personalized virtual heart simulations faces challenges in model personalization and expensive computation. While an ideal solution is an efficient neural surrogate that at the same time is personalized to an individual subject, the state-of-the-art is either concerned with personalizing an expensive simulation model, or learning an efficient yet generic surrogate. This paper presents a completely new concept to achieve personalized neural surrogates in a single coherent framework of meta-learning (metaPNS). Instead of learning a single neural surrogate, we pursue the process of learning a personalized neural surrogate using a small amount of context data from a subject, in a novel formulation of few-shot generative modeling underpinned by: 1) a set-conditioned neural surrogate for cardiac simulation that, conditioned on subject-specific context data, learns to generate query simulations not included in the context set, and 2) a meta-model of amortized variational inference that learns to condition the neural surrogate via simple feed-forward embedding of context data. As test time, metaPNS delivers a personalized neural surrogate by fast feed-forward embedding of a small and flexible number of data available from an individual, achieving -- for the first time -- personalization and surrogate construction for expensive simulations in one end-to-end learning framework. Synthetic and real-data experiments demonstrated that metaPNS was able to improve personalization and predictive accuracy in comparison to conventionally-optimized cardiac simulation models, at a fraction of computation.
Neural State-Space Modeling with Latent Causal-Effect Disentanglement
Sep 26, 2022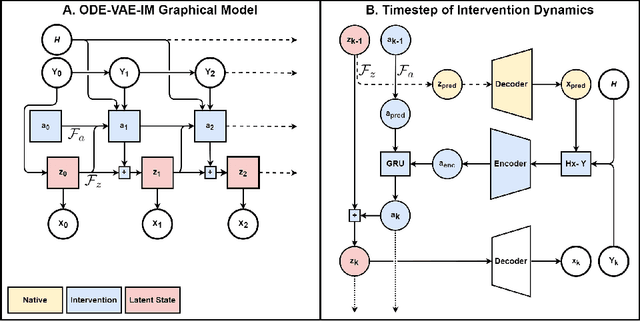
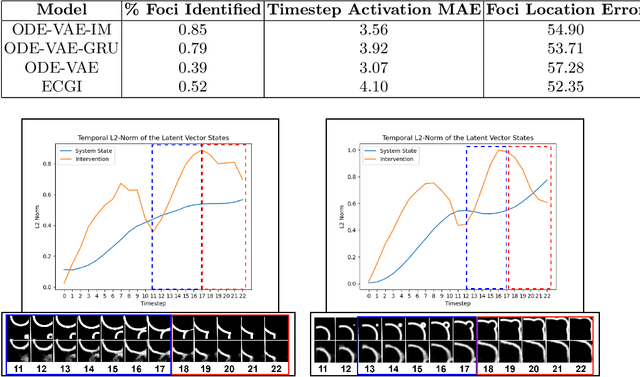
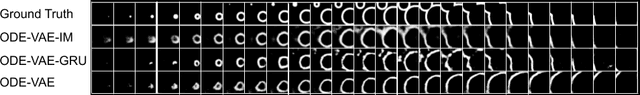
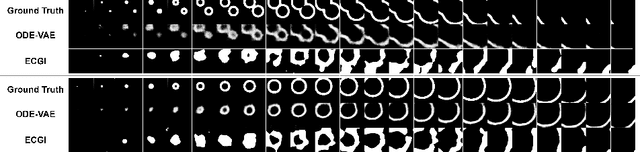
Abstract:Despite substantial progress in deep learning approaches to time-series reconstruction, no existing methods are designed to uncover local activities with minute signal strength due to their negligible contribution to the optimization loss. Such local activities however can signify important abnormal events in physiological systems, such as an extra foci triggering an abnormal propagation of electrical waves in the heart. We discuss a novel technique for reconstructing such local activity that, while small in signal strength, is the cause of subsequent global activities that have larger signal strength. Our central innovation is to approach this by explicitly modeling and disentangling how the latent state of a system is influenced by potential hidden internal interventions. In a novel neural formulation of state-space models (SSMs), we first introduce causal-effect modeling of the latent dynamics via a system of interacting neural ODEs that separately describes 1) the continuous-time dynamics of the internal intervention, and 2) its effect on the trajectory of the system's native state. Because the intervention can not be directly observed but have to be disentangled from the observed subsequent effect, we integrate knowledge of the native intervention-free dynamics of a system, and infer the hidden intervention by assuming it to be responsible for differences observed between the actual and hypothetical intervention-free dynamics. We demonstrated a proof-of-concept of the presented framework on reconstructing ectopic foci disrupting the course of normal cardiac electrical propagation from remote observations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge