Yubo Ye
HyPer-EP: Meta-Learning Hybrid Personalized Models for Cardiac Electrophysiology
Mar 15, 2024


Abstract:Personalized virtual heart models have demonstrated increasing potential for clinical use, although the estimation of their parameters given patient-specific data remain a challenge. Traditional physics-based modeling approaches are computationally costly and often neglect the inherent structural errors in these models due to model simplifications and assumptions. Modern deep learning approaches, on the other hand, rely heavily on data supervision and lacks interpretability. In this paper, we present a novel hybrid modeling framework to describe a personalized cardiac digital twin as a combination of a physics-based known expression augmented by neural network modeling of its unknown gap to reality. We then present a novel meta-learning framework to enable the separate identification of both the physics-based and neural components in the hybrid model. We demonstrate the feasibility and generality of this hybrid modeling framework with two examples of instantiations and their proof-of-concept in synthetic experiments.
Unsupervised Learning of Hybrid Latent Dynamics: A Learn-to-Identify Framework
Mar 13, 2024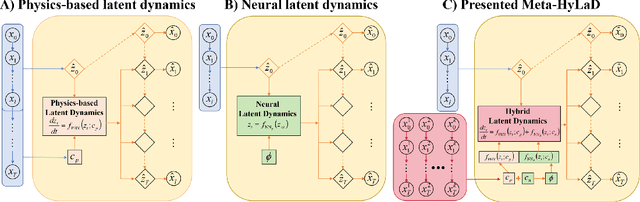
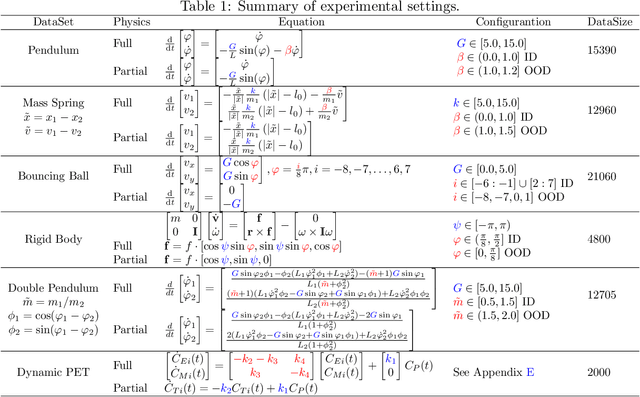
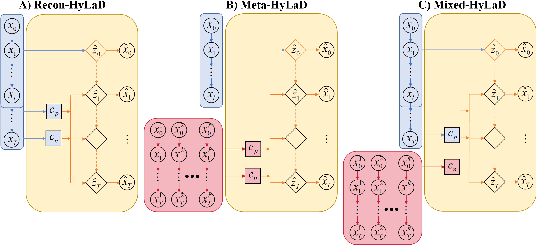
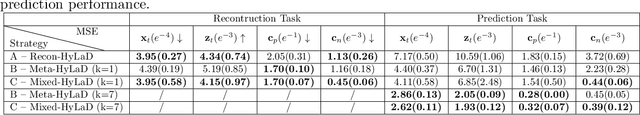
Abstract:Modern applications increasingly require unsupervised learning of latent dynamics from high-dimensional time-series. This presents a significant challenge of identifiability: many abstract latent representations may reconstruct observations, yet do they guarantee an adequate identification of the governing dynamics? This paper investigates this challenge from two angles: the use of physics inductive bias specific to the data being modeled, and a learn-to-identify strategy that separates forecasting objectives from the data used for the identification. We combine these two strategies in a novel framework for unsupervised meta-learning of hybrid latent dynamics (Meta-HyLaD) with: 1) a latent dynamic function that hybridize known mathematical expressions of prior physics with neural functions describing its unknown errors, and 2) a meta-learning formulation to learn to separately identify both components of the hybrid dynamics. Through extensive experiments on five physics and one biomedical systems, we provide strong evidence for the benefits of Meta-HyLaD to integrate rich prior knowledge while identifying their gap to observed data.
Hybrid Kinetics Embedding Framework for Dynamic PET Reconstruction
Mar 12, 2024Abstract:In dynamic positron emission tomography (PET) reconstruction, the importance of leveraging the temporal dependence of the data has been well appreciated. Current deep-learning solutions can be categorized in two groups in the way the temporal dynamics is modeled: data-driven approaches use spatiotemporal neural networks to learn the temporal dynamics of tracer kinetics from data, which relies heavily on data supervision; physics-based approaches leverage \textit{a priori} tracer kinetic models to focus on inferring their parameters, which relies heavily on the accuracy of the prior kinetic model. In this paper, we marry the strengths of these two approaches in a hybrid kinetics embedding (HyKE-Net) framework for dynamic PET reconstruction. We first introduce a novel \textit{hybrid} model of tracer kinetics consisting of a physics-based function augmented by a neural component to account for its gap to data-generating tracer kinetics, both identifiable from data. We then embed this hybrid model at the latent space of an encoding-decoding framework to enable both supervised and unsupervised identification of the hybrid kinetics and thereby dynamic PET reconstruction. Through both phantom and real-data experiments, we demonstrate the benefits of HyKE-Net -- especially in unsupervised reconstructions -- over existing physics-based and data-driven baselines as well as its ablated formulations where the embedded tracer kinetics are purely physics-based, purely neural, or hybrid but with a non-adaptable neural component.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge