Lianming Wu
A3-TTA: Adaptive Anchor Alignment Test-Time Adaptation for Image Segmentation
Feb 03, 2026Abstract:Test-Time Adaptation (TTA) offers a practical solution for deploying image segmentation models under domain shift without accessing source data or retraining. Among existing TTA strategies, pseudo-label-based methods have shown promising performance. However, they often rely on perturbation-ensemble heuristics (e.g., dropout sampling, test-time augmentation, Gaussian noise), which lack distributional grounding and yield unstable training signals. This can trigger error accumulation and catastrophic forgetting during adaptation. To address this, we propose \textbf{A3-TTA}, a TTA framework that constructs reliable pseudo-labels through anchor-guided supervision. Specifically, we identify well-predicted target domain images using a class compact density metric, under the assumption that confident predictions imply distributional proximity to the source domain. These anchors serve as stable references to guide pseudo-label generation, which is further regularized via semantic consistency and boundary-aware entropy minimization. Additionally, we introduce a self-adaptive exponential moving average strategy to mitigate label noise and stabilize model update during adaptation. Evaluated on both multi-domain medical images (heart structure and prostate segmentation) and natural images, A3-TTA significantly improves average Dice scores by 10.40 to 17.68 percentage points compared to the source model, outperforming several state-of-the-art TTA methods under different segmentation model architectures. A3-TTA also excels in continual TTA, maintaining high performance across sequential target domains with strong anti-forgetting ability. The code will be made publicly available at https://github.com/HiLab-git/A3-TTA.
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
CTSL: Codebook-based Temporal-Spatial Learning for Accurate Non-Contrast Cardiac Risk Prediction Using Cine MRIs
Jul 22, 2025Abstract:Accurate and contrast-free Major Adverse Cardiac Events (MACE) prediction from Cine MRI sequences remains a critical challenge. Existing methods typically necessitate supervised learning based on human-refined masks in the ventricular myocardium, which become impractical without contrast agents. We introduce a self-supervised framework, namely Codebook-based Temporal-Spatial Learning (CTSL), that learns dynamic, spatiotemporal representations from raw Cine data without requiring segmentation masks. CTSL decouples temporal and spatial features through a multi-view distillation strategy, where the teacher model processes multiple Cine views, and the student model learns from reduced-dimensional Cine-SA sequences. By leveraging codebook-based feature representations and dynamic lesion self-detection through motion cues, CTSL captures intricate temporal dependencies and motion patterns. High-confidence MACE risk predictions are achieved through our model, providing a rapid, non-invasive solution for cardiac risk assessment that outperforms traditional contrast-dependent methods, thereby enabling timely and accessible heart disease diagnosis in clinical settings.
A Composite Alignment-Aware Framework for Myocardial Lesion Segmentation in Multi-sequence CMR Images
Jul 16, 2025Abstract:Accurate segmentation of myocardial lesions from multi-sequence cardiac magnetic resonance imaging is essential for cardiac disease diagnosis and treatment planning. However, achieving optimal feature correspondence is challenging due to intensity variations across modalities and spatial misalignment caused by inconsistent slice acquisition protocols. We propose CAA-Seg, a composite alignment-aware framework that addresses these challenges through a two-stage approach. First, we introduce a selective slice alignment method that dynamically identifies and aligns anatomically corresponding slice pairs while excluding mismatched sections, ensuring reliable spatial correspondence between sequences. Second, we develop a hierarchical alignment network that processes multi-sequence features at different semantic levels, i.e., local deformation correction modules address geometric variations in low-level features, while global semantic fusion blocks enable semantic fusion at high levels where intensity discrepancies diminish. We validate our method on a large-scale dataset comprising 397 patients. Experimental results show that our proposed CAA-Seg achieves superior performance on most evaluation metrics, with particularly strong results in myocardial infarction segmentation, representing a substantial 5.54% improvement over state-of-the-art approaches. The code is available at https://github.com/yifangao112/CAA-Seg.
CineMyoPS: Segmenting Myocardial Pathologies from Cine Cardiac MR
Jul 03, 2025



Abstract:Myocardial infarction (MI) is a leading cause of death worldwide. Late gadolinium enhancement (LGE) and T2-weighted cardiac magnetic resonance (CMR) imaging can respectively identify scarring and edema areas, both of which are essential for MI risk stratification and prognosis assessment. Although combining complementary information from multi-sequence CMR is useful, acquiring these sequences can be time-consuming and prohibitive, e.g., due to the administration of contrast agents. Cine CMR is a rapid and contrast-free imaging technique that can visualize both motion and structural abnormalities of the myocardium induced by acute MI. Therefore, we present a new end-to-end deep neural network, referred to as CineMyoPS, to segment myocardial pathologies, \ie scars and edema, solely from cine CMR images. Specifically, CineMyoPS extracts both motion and anatomy features associated with MI. Given the interdependence between these features, we design a consistency loss (resembling the co-training strategy) to facilitate their joint learning. Furthermore, we propose a time-series aggregation strategy to integrate MI-related features across the cardiac cycle, thereby enhancing segmentation accuracy for myocardial pathologies. Experimental results on a multi-center dataset demonstrate that CineMyoPS achieves promising performance in myocardial pathology segmentation, motion estimation, and anatomy segmentation.
U2AD: Uncertainty-based Unsupervised Anomaly Detection Framework for Detecting T2 Hyperintensity in MRI Spinal Cord
Mar 17, 2025Abstract:T2 hyperintensities in spinal cord MR images are crucial biomarkers for conditions such as degenerative cervical myelopathy. However, current clinical diagnoses primarily rely on manual evaluation. Deep learning methods have shown promise in lesion detection, but most supervised approaches are heavily dependent on large, annotated datasets. Unsupervised anomaly detection (UAD) offers a compelling alternative by eliminating the need for abnormal data annotations. However, existing UAD methods rely on curated normal datasets and their performance frequently deteriorates when applied to clinical datasets due to domain shifts. We propose an Uncertainty-based Unsupervised Anomaly Detection framework, termed U2AD, to address these limitations. Unlike traditional methods, U2AD is designed to be trained and tested within the same clinical dataset, following a "mask-and-reconstruction" paradigm built on a Vision Transformer-based architecture. We introduce an uncertainty-guided masking strategy to resolve task conflicts between normal reconstruction and anomaly detection to achieve an optimal balance. Specifically, we employ a Monte-Carlo sampling technique to estimate reconstruction uncertainty mappings during training. By iteratively optimizing reconstruction training under the guidance of both epistemic and aleatoric uncertainty, U2AD reduces overall reconstruction variance while emphasizing regions. Experimental results demonstrate that U2AD outperforms existing supervised and unsupervised methods in patient-level identification and segment-level localization tasks. This framework establishes a new benchmark for incorporating uncertainty guidance into UAD, highlighting its clinical utility in addressing domain shifts and task conflicts in medical image anomaly detection. Our code is available: https://github.com/zhibaishouheilab/U2AD
Pathology-Guided AI System for Accurate Segmentation and Diagnosis of Cervical Spondylosis
Mar 08, 2025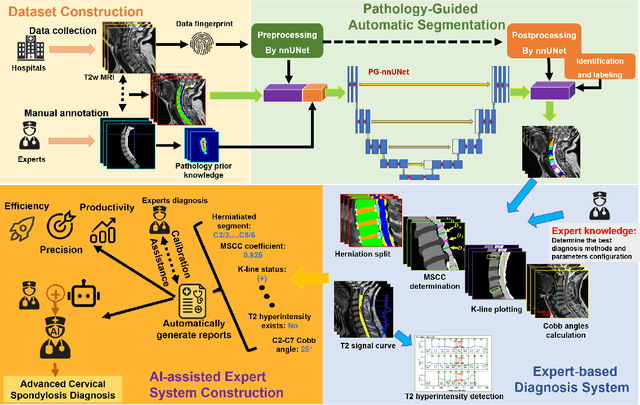
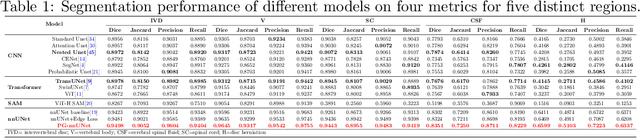
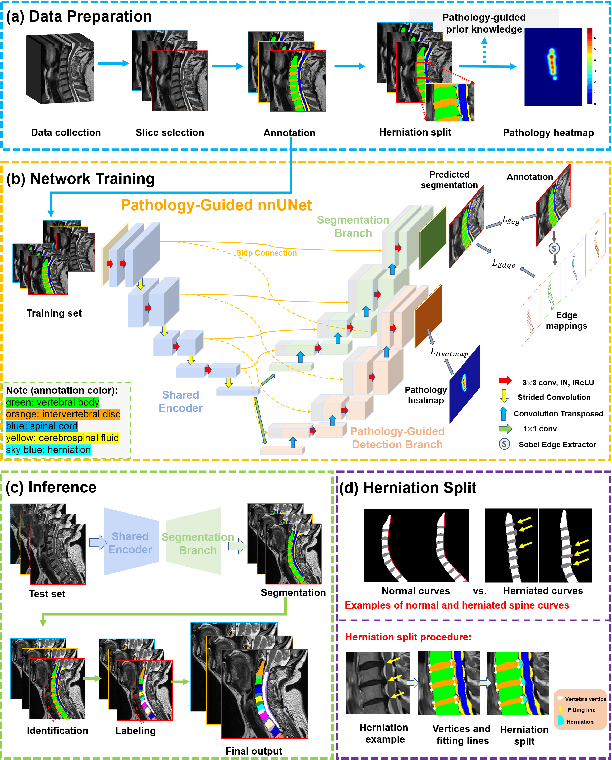
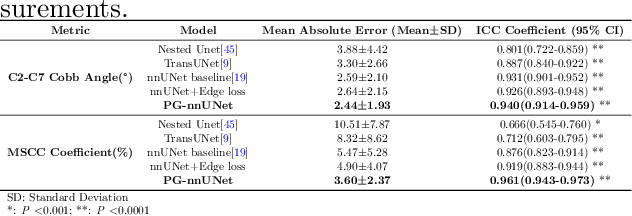
Abstract:Cervical spondylosis, a complex and prevalent condition, demands precise and efficient diagnostic techniques for accurate assessment. While MRI offers detailed visualization of cervical spine anatomy, manual interpretation remains labor-intensive and prone to error. To address this, we developed an innovative AI-assisted Expert-based Diagnosis System that automates both segmentation and diagnosis of cervical spondylosis using MRI. Leveraging a dataset of 960 cervical MRI images from patients with cervical disc herniation, our system features a pathology-guided segmentation model capable of accurately segmenting key cervical anatomical structures. The segmentation is followed by an expert-based diagnostic framework that automates the calculation of critical clinical indicators. Our segmentation model achieved an impressive average Dice coefficient exceeding 0.90 across four cervical spinal anatomies and demonstrated enhanced accuracy in herniation areas. Diagnostic evaluation further showcased the system precision, with a mean absolute error (MAE) of 2.44 degree for the C2-C7 Cobb angle and 3.60 precentage for the Maximum Spinal Cord Compression (MSCC) coefficient. In addition, our method delivered high accuracy, precision, recall, and F1 scores in herniation localization, K-line status assessment, and T2 hyperintensity detection. Comparative analysis demonstrates that our system outperforms existing methods, establishing a new benchmark for segmentation and diagnostic tasks for cervical spondylosis.
CMRxRecon2024: A Multi-Modality, Multi-View K-Space Dataset Boosting Universal Machine Learning for Accelerated Cardiac MRI
Jun 27, 2024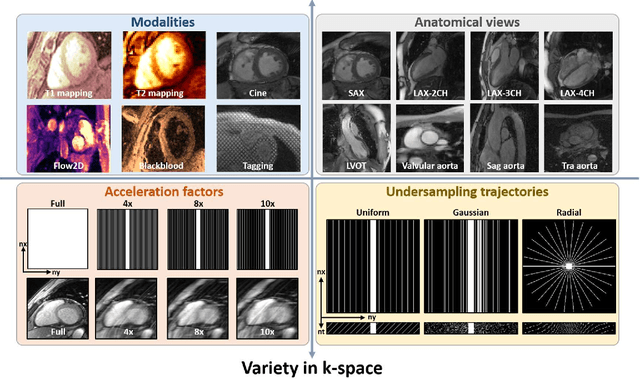
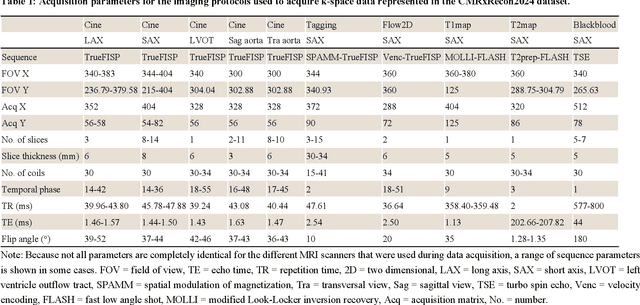
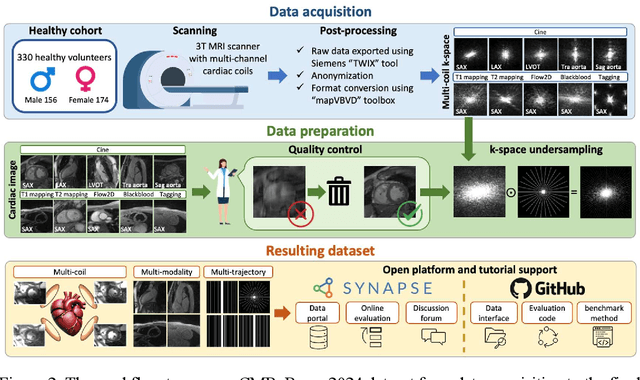
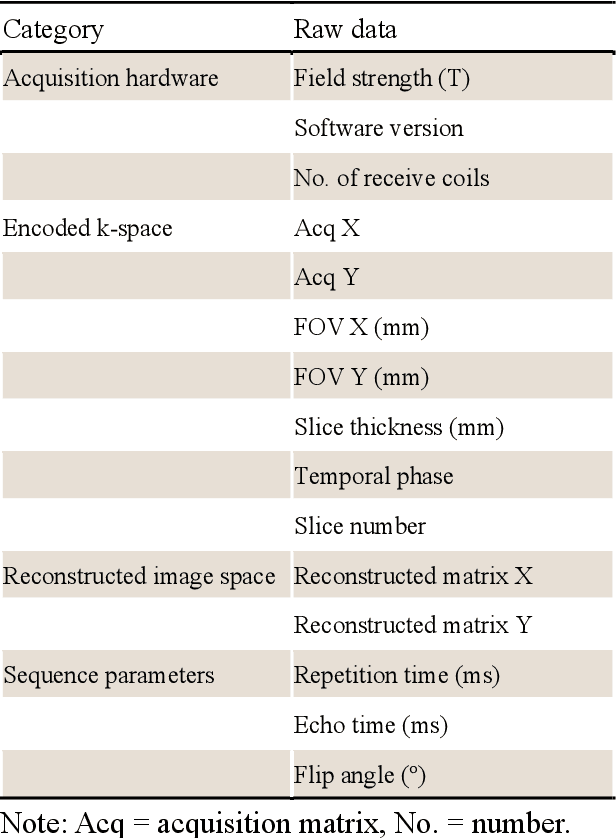
Abstract:Cardiac magnetic resonance imaging (MRI) has emerged as a clinically gold-standard technique for diagnosing cardiac diseases, thanks to its ability to provide diverse information with multiple modalities and anatomical views. Accelerated cardiac MRI is highly expected to achieve time-efficient and patient-friendly imaging, and then advanced image reconstruction approaches are required to recover high-quality, clinically interpretable images from undersampled measurements. However, the lack of publicly available cardiac MRI k-space dataset in terms of both quantity and diversity has severely hindered substantial technological progress, particularly for data-driven artificial intelligence. Here, we provide a standardized, diverse, and high-quality CMRxRecon2024 dataset to facilitate the technical development, fair evaluation, and clinical transfer of cardiac MRI reconstruction approaches, towards promoting the universal frameworks that enable fast and robust reconstructions across different cardiac MRI protocols in clinical practice. To the best of our knowledge, the CMRxRecon2024 dataset is the largest and most diverse publicly available cardiac k-space dataset. It is acquired from 330 healthy volunteers, covering commonly used modalities, anatomical views, and acquisition trajectories in clinical cardiac MRI workflows. Besides, an open platform with tutorials, benchmarks, and data processing tools is provided to facilitate data usage, advanced method development, and fair performance evaluation.
Deep learning-driven pulmonary arteries and veins segmentation reveals demography-associated pulmonary vasculature anatomy
Apr 11, 2024Abstract:Pulmonary artery-vein segmentation is crucial for diagnosing pulmonary diseases and surgical planning, and is traditionally achieved by Computed Tomography Pulmonary Angiography (CTPA). However, concerns regarding adverse health effects from contrast agents used in CTPA have constrained its clinical utility. In contrast, identifying arteries and veins using non-contrast CT, a conventional and low-cost clinical examination routine, has long been considered impossible. Here we propose a High-abundant Pulmonary Artery-vein Segmentation (HiPaS) framework achieving accurate artery-vein segmentation on both non-contrast CT and CTPA across various spatial resolutions. HiPaS first performs spatial normalization on raw CT scans via a super-resolution module, and then iteratively achieves segmentation results at different branch levels by utilizing the low-level vessel segmentation as a prior for high-level vessel segmentation. We trained and validated HiPaS on our established multi-centric dataset comprising 1,073 CT volumes with meticulous manual annotation. Both quantitative experiments and clinical evaluation demonstrated the superior performance of HiPaS, achieving a dice score of 91.8% and a sensitivity of 98.0%. Further experiments demonstrated the non-inferiority of HiPaS segmentation on non-contrast CT compared to segmentation on CTPA. Employing HiPaS, we have conducted an anatomical study of pulmonary vasculature on 10,613 participants in China (five sites), discovering a new association between pulmonary vessel abundance and sex and age: vessel abundance is significantly higher in females than in males, and slightly decreases with age, under the controlling of lung volumes (p < 0.0001). HiPaS realizing accurate artery-vein segmentation delineates a promising avenue for clinical diagnosis and understanding pulmonary physiology in a non-invasive manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge