Kaiwen Dong
RIMRULE: Improving Tool-Using Language Agents via MDL-Guided Rule Learning
Jan 05, 2026Abstract:Large language models (LLMs) often struggle to use tools reliably in domain-specific settings, where APIs may be idiosyncratic, under-documented, or tailored to private workflows. This highlights the need for effective adaptation to task-specific tools. We propose RIMRULE, a neuro-symbolic approach for LLM adaptation based on dynamic rule injection. Compact, interpretable rules are distilled from failure traces and injected into the prompt during inference to improve task performance. These rules are proposed by the LLM itself and consolidated using a Minimum Description Length (MDL) objective that favors generality and conciseness. Each rule is stored in both natural language and a structured symbolic form, supporting efficient retrieval at inference time. Experiments on tool-use benchmarks show that this approach improves accuracy on both seen and unseen tools without modifying LLM weights. It outperforms prompting-based adaptation methods and complements finetuning. Moreover, rules learned from one LLM can be reused to improve others, including long reasoning LLMs, highlighting the portability of symbolic knowledge across architectures.
MolX: Enhancing Large Language Models for Molecular Learning with A Multi-Modal Extension
Jun 10, 2024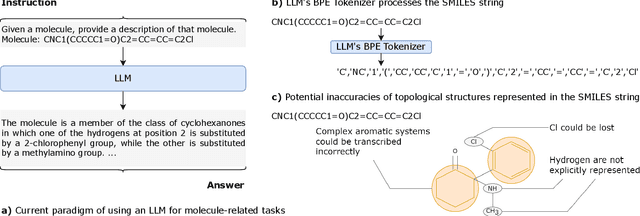
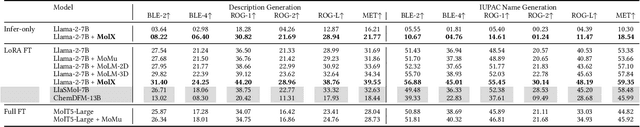
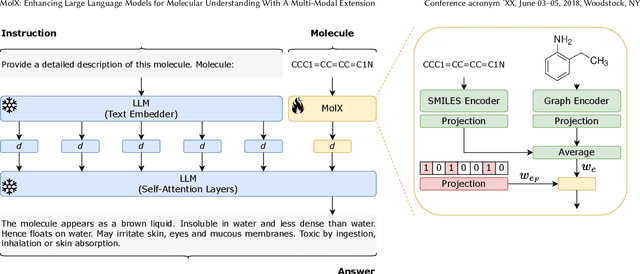
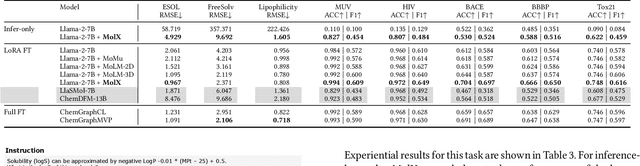
Abstract:Recently, Large Language Models (LLMs) with their strong task-handling capabilities have shown remarkable advancements across a spectrum of fields, moving beyond natural language understanding. However, their proficiency within the chemistry domain remains restricted, especially in solving professional molecule-related tasks. This challenge is attributed to their inherent limitations in comprehending molecules using only common textual representations, i.e., SMILES strings. In this study, we seek to enhance the ability of LLMs to comprehend molecules by designing and equipping them with a multi-modal external module, namely MolX. In particular, instead of directly using a SMILES string to represent a molecule, we utilize specific encoders to extract fine-grained features from both SMILES string and 2D molecular graph representations for feeding into an LLM. Moreover, a human-defined molecular fingerprint is incorporated to leverage its embedded domain knowledge. Then, to establish an alignment between MolX and the LLM's textual input space, the whole model in which the LLM is frozen, is pre-trained with a versatile strategy including a diverse set of tasks. Extensive experimental evaluations demonstrate that our proposed method only introduces a small number of trainable parameters while outperforming baselines on various downstream molecule-related tasks ranging from molecule-to-text translation to retrosynthesis, with and without fine-tuning the LLM.
You do not have to train Graph Neural Networks at all on text-attributed graphs
Apr 17, 2024
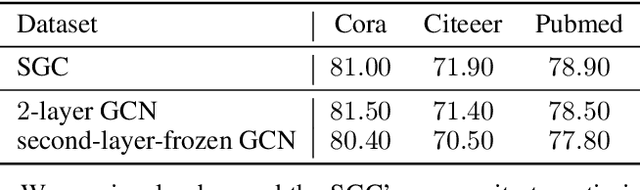
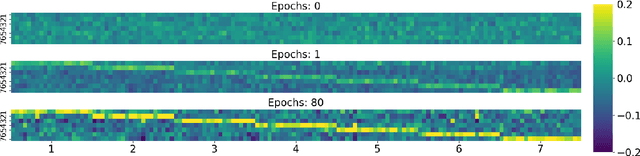
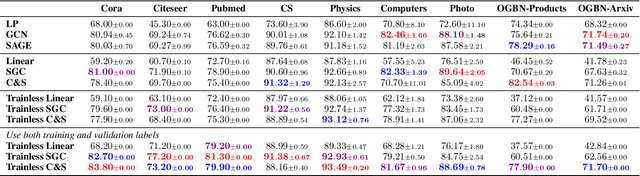
Abstract:Graph structured data, specifically text-attributed graphs (TAG), effectively represent relationships among varied entities. Such graphs are essential for semi-supervised node classification tasks. Graph Neural Networks (GNNs) have emerged as a powerful tool for handling this graph-structured data. Although gradient descent is commonly utilized for training GNNs for node classification, this study ventures into alternative methods, eliminating the iterative optimization processes. We introduce TrainlessGNN, a linear GNN model capitalizing on the observation that text encodings from the same class often cluster together in a linear subspace. This model constructs a weight matrix to represent each class's node attribute subspace, offering an efficient approach to semi-supervised node classification on TAG. Extensive experiments reveal that our trainless models can either match or even surpass their conventionally trained counterparts, demonstrating the possibility of refraining from gradient descent in certain configurations.
CORE: Data Augmentation for Link Prediction via Information Bottleneck
Apr 17, 2024
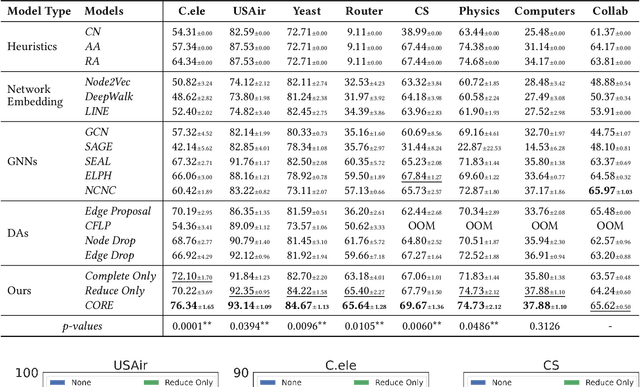

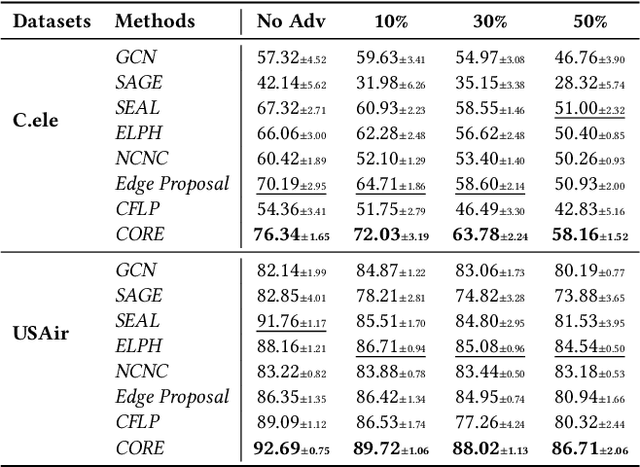
Abstract:Link prediction (LP) is a fundamental task in graph representation learning, with numerous applications in diverse domains. However, the generalizability of LP models is often compromised due to the presence of noisy or spurious information in graphs and the inherent incompleteness of graph data. To address these challenges, we draw inspiration from the Information Bottleneck principle and propose a novel data augmentation method, COmplete and REduce (CORE) to learn compact and predictive augmentations for LP models. In particular, CORE aims to recover missing edges in graphs while simultaneously removing noise from the graph structures, thereby enhancing the model's robustness and performance. Extensive experiments on multiple benchmark datasets demonstrate the applicability and superiority of CORE over state-of-the-art methods, showcasing its potential as a leading approach for robust LP in graph representation learning.
Node Duplication Improves Cold-start Link Prediction
Feb 15, 2024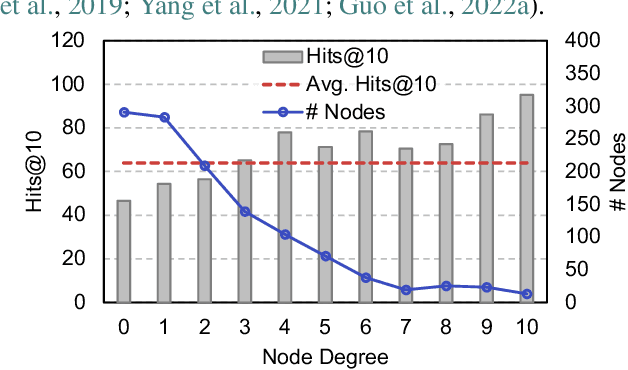
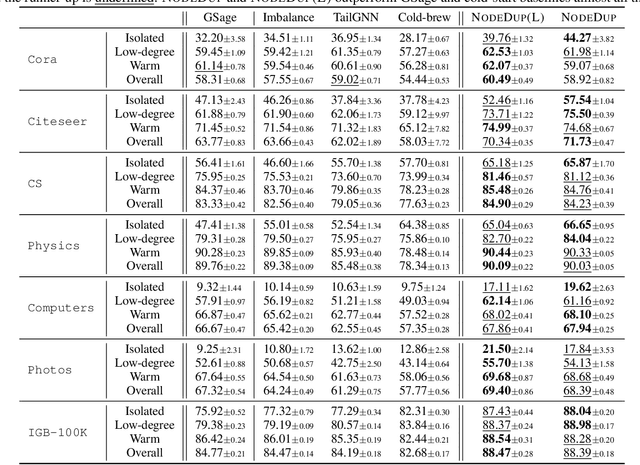
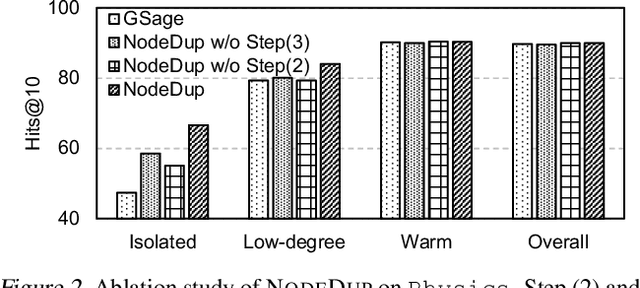
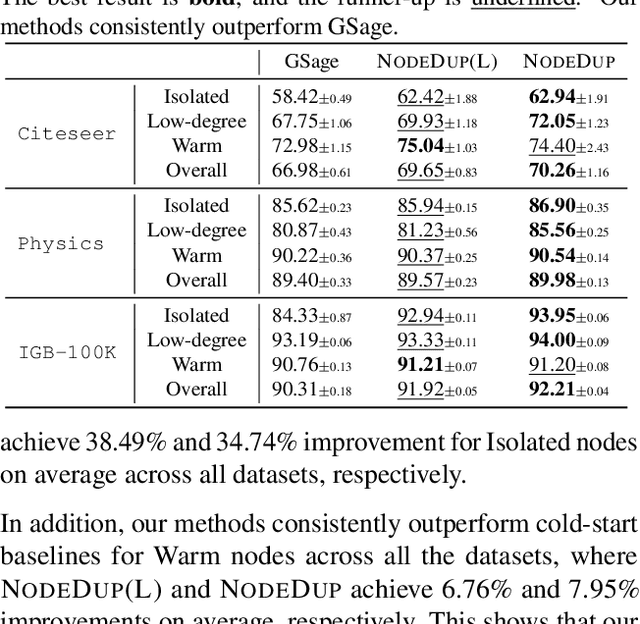
Abstract:Graph Neural Networks (GNNs) are prominent in graph machine learning and have shown state-of-the-art performance in Link Prediction (LP) tasks. Nonetheless, recent studies show that GNNs struggle to produce good results on low-degree nodes despite their overall strong performance. In practical applications of LP, like recommendation systems, improving performance on low-degree nodes is critical, as it amounts to tackling the cold-start problem of improving the experiences of users with few observed interactions. In this paper, we investigate improving GNNs' LP performance on low-degree nodes while preserving their performance on high-degree nodes and propose a simple yet surprisingly effective augmentation technique called NodeDup. Specifically, NodeDup duplicates low-degree nodes and creates links between nodes and their own duplicates before following the standard supervised LP training scheme. By leveraging a ''multi-view'' perspective for low-degree nodes, NodeDup shows significant LP performance improvements on low-degree nodes without compromising any performance on high-degree nodes. Additionally, as a plug-and-play augmentation module, NodeDup can be easily applied to existing GNNs with very light computational cost. Extensive experiments show that NodeDup achieves 38.49%, 13.34%, and 6.76% improvements on isolated, low-degree, and warm nodes, respectively, on average across all datasets compared to GNNs and state-of-the-art cold-start methods.
Universal Link Predictor By In-Context Learning on Graphs
Feb 15, 2024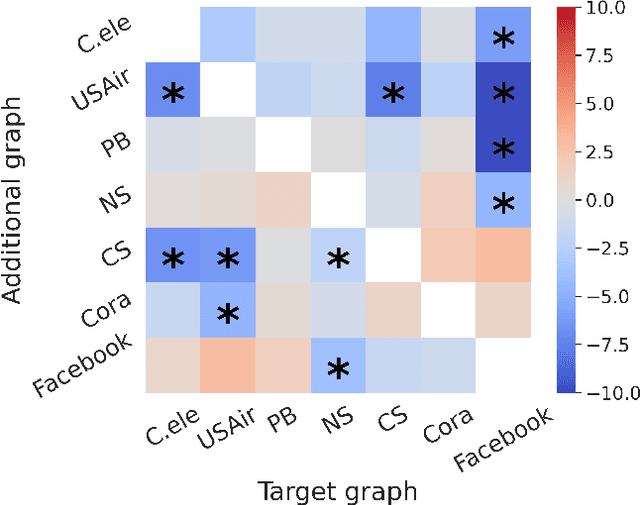
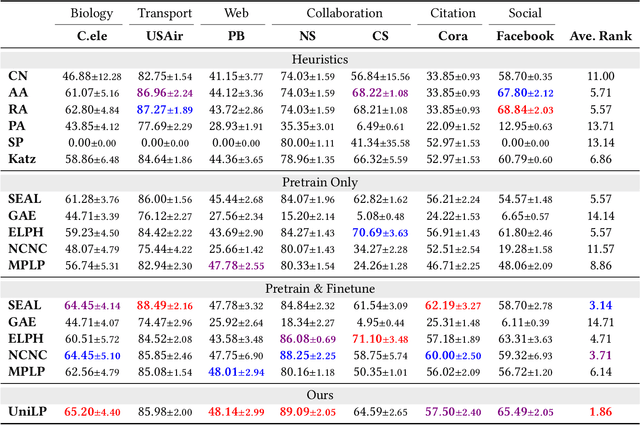
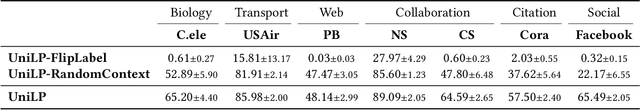
Abstract:Link prediction is a crucial task in graph machine learning, where the goal is to infer missing or future links within a graph. Traditional approaches leverage heuristic methods based on widely observed connectivity patterns, offering broad applicability and generalizability without the need for model training. Despite their utility, these methods are limited by their reliance on human-derived heuristics and lack the adaptability of data-driven approaches. Conversely, parametric link predictors excel in automatically learning the connectivity patterns from data and achieving state-of-the-art but fail short to directly transfer across different graphs. Instead, it requires the cost of extensive training and hyperparameter optimization to adapt to the target graph. In this work, we introduce the Universal Link Predictor (UniLP), a novel model that combines the generalizability of heuristic approaches with the pattern learning capabilities of parametric models. UniLP is designed to autonomously identify connectivity patterns across diverse graphs, ready for immediate application to any unseen graph dataset without targeted training. We address the challenge of conflicting connectivity patterns-arising from the unique distributions of different graphs-through the implementation of In-context Learning (ICL). This approach allows UniLP to dynamically adjust to various target graphs based on contextual demonstrations, thereby avoiding negative transfer. Through rigorous experimentation, we demonstrate UniLP's effectiveness in adapting to new, unseen graphs at test time, showcasing its ability to perform comparably or even outperform parametric models that have been finetuned for specific datasets. Our findings highlight UniLP's potential to set a new standard in link prediction, combining the strengths of heuristic and parametric methods in a single, versatile framework.
Pure Message Passing Can Estimate Common Neighbor for Link Prediction
Sep 02, 2023



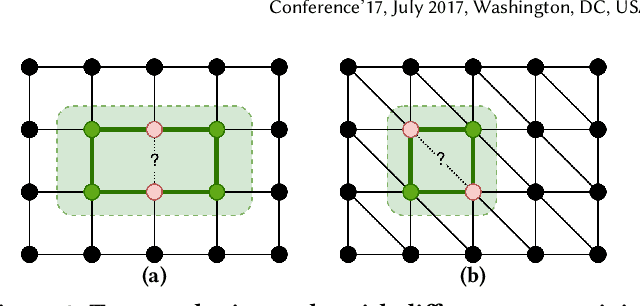
Abstract:Message Passing Neural Networks (MPNNs) have emerged as the {\em de facto} standard in graph representation learning. However, when it comes to link prediction, they often struggle, surpassed by simple heuristics such as Common Neighbor (CN). This discrepancy stems from a fundamental limitation: while MPNNs excel in node-level representation, they stumble with encoding the joint structural features essential to link prediction, like CN. To bridge this gap, we posit that, by harnessing the orthogonality of input vectors, pure message-passing can indeed capture joint structural features. Specifically, we study the proficiency of MPNNs in approximating CN heuristics. Based on our findings, we introduce the Message Passing Link Predictor (MPLP), a novel link prediction model. MPLP taps into quasi-orthogonal vectors to estimate link-level structural features, all while preserving the node-level complexities. Moreover, our approach demonstrates that leveraging message-passing to capture structural features could offset MPNNs' expressiveness limitations at the expense of estimation variance. We conduct experiments on benchmark datasets from various domains, where our method consistently outperforms the baseline methods.
FakeEdge: Alleviate Dataset Shift in Link Prediction
Dec 03, 2022



Abstract:Link prediction is a crucial problem in graph-structured data. Due to the recent success of graph neural networks (GNNs), a variety of GNN-based models were proposed to tackle the link prediction task. Specifically, GNNs leverage the message passing paradigm to obtain node representation, which relies on link connectivity. However, in a link prediction task, links in the training set are always present while ones in the testing set are not yet formed, resulting in a discrepancy of the connectivity pattern and bias of the learned representation. It leads to a problem of dataset shift which degrades the model performance. In this paper, we first identify the dataset shift problem in the link prediction task and provide theoretical analyses on how existing link prediction methods are vulnerable to it. We then propose FakeEdge, a model-agnostic technique, to address the problem by mitigating the graph topological gap between training and testing sets. Extensive experiments demonstrate the applicability and superiority of FakeEdge on multiple datasets across various domains.
Heterogeneous Graph Masked Autoencoders
Aug 21, 2022



Abstract:Generative self-supervised learning (SSL), especially masked autoencoders, has become one of the most exciting learning paradigms and has shown great potential in handling graph data. However, real-world graphs are always heterogeneous, which poses three critical challenges that existing methods ignore: 1) how to capture complex graph structure? 2) how to incorporate various node attributes? and 3) how to encode different node positions? In light of this, we study the problem of generative SSL on heterogeneous graphs and propose HGMAE, a novel heterogeneous graph masked autoencoder model to address these challenges. HGMAE captures comprehensive graph information via two innovative masking techniques and three unique training strategies. In particular, we first develop metapath masking and adaptive attribute masking with dynamic mask rate to enable effective and stable learning on heterogeneous graphs. We then design several training strategies including metapath-based edge reconstruction to adopt complex structural information, target attribute restoration to incorporate various node attributes, and positional feature prediction to encode node positional information. Extensive experiments demonstrate that HGMAE outperforms both contrastive and generative state-of-the-art baselines on several tasks across multiple datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge