Xiaobao Huang
CrochetBench: Can Vision-Language Models Move from Describing to Doing in Crochet Domain?
Nov 12, 2025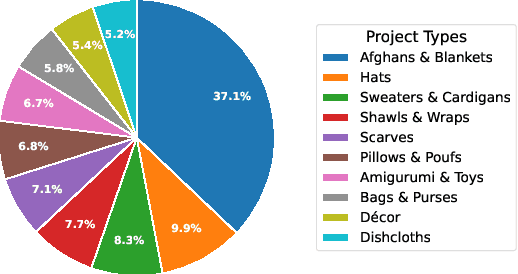

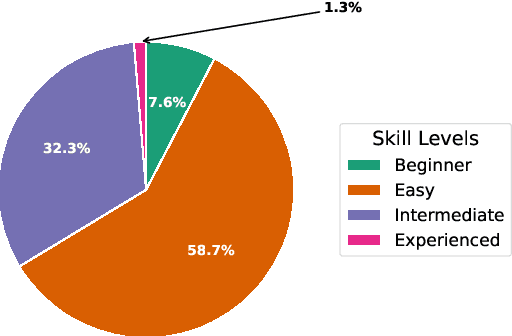
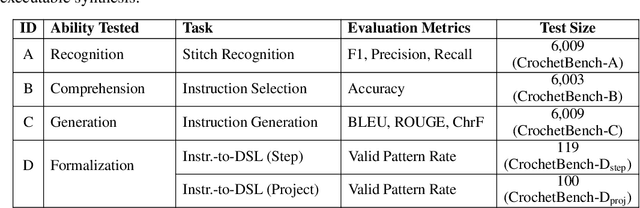
Abstract:We present CrochetBench, a benchmark for evaluating the ability of multimodal large language models to perform fine-grained, low-level procedural reasoning in the domain of crochet. Unlike prior benchmarks that focus on high-level description or visual question answering, CrochetBench shifts the emphasis from describing to doing: models are required to recognize stitches, select structurally appropriate instructions, and generate compilable crochet procedures. We adopt the CrochetPARADE DSL as our intermediate representation, enabling structural validation and functional evaluation via execution. The benchmark covers tasks including stitch classification, instruction grounding, and both natural language and image-to-DSL translation. Across all tasks, performance sharply declines as the evaluation shifts from surface-level similarity to executable correctness, exposing limitations in long-range symbolic reasoning and 3D-aware procedural synthesis. CrochetBench offers a new lens for assessing procedural competence in multimodal models and highlights the gap between surface-level understanding and executable precision in real-world creative domains. Code is available at https://github.com/Peiyu-Georgia-Li/crochetBench.
ChefFusion: Multimodal Foundation Model Integrating Recipe and Food Image Generation
Sep 18, 2024


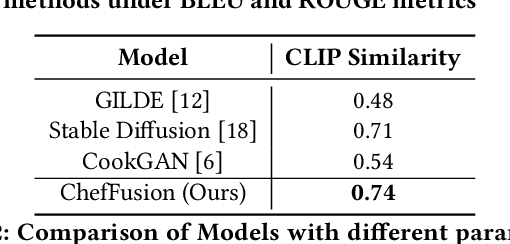
Abstract:Significant work has been conducted in the domain of food computing, yet these studies typically focus on single tasks such as t2t (instruction generation from food titles and ingredients), i2t (recipe generation from food images), or t2i (food image generation from recipes). None of these approaches integrate all modalities simultaneously. To address this gap, we introduce a novel food computing foundation model that achieves true multimodality, encompassing tasks such as t2t, t2i, i2t, it2t, and t2ti. By leveraging large language models (LLMs) and pre-trained image encoder and decoder models, our model can perform a diverse array of food computing-related tasks, including food understanding, food recognition, recipe generation, and food image generation. Compared to previous models, our foundation model demonstrates a significantly broader range of capabilities and exhibits superior performance, particularly in food image generation and recipe generation tasks. We open-sourced ChefFusion at GitHub.
MolX: Enhancing Large Language Models for Molecular Learning with A Multi-Modal Extension
Jun 10, 2024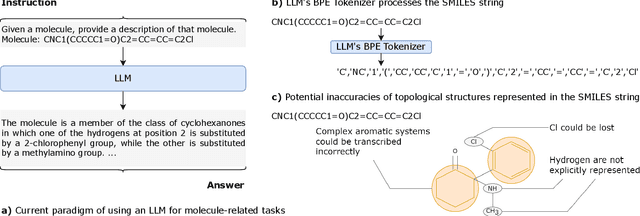
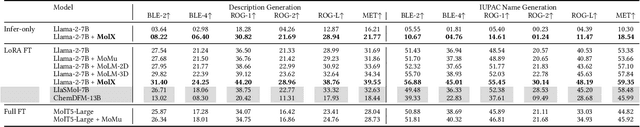
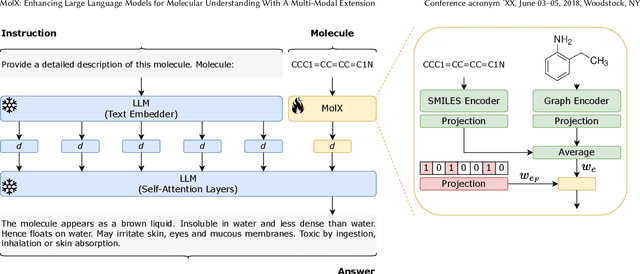
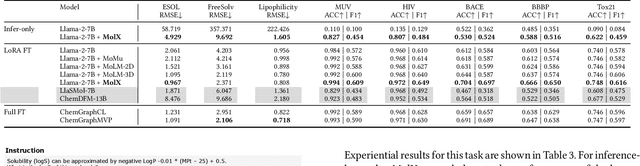
Abstract:Recently, Large Language Models (LLMs) with their strong task-handling capabilities have shown remarkable advancements across a spectrum of fields, moving beyond natural language understanding. However, their proficiency within the chemistry domain remains restricted, especially in solving professional molecule-related tasks. This challenge is attributed to their inherent limitations in comprehending molecules using only common textual representations, i.e., SMILES strings. In this study, we seek to enhance the ability of LLMs to comprehend molecules by designing and equipping them with a multi-modal external module, namely MolX. In particular, instead of directly using a SMILES string to represent a molecule, we utilize specific encoders to extract fine-grained features from both SMILES string and 2D molecular graph representations for feeding into an LLM. Moreover, a human-defined molecular fingerprint is incorporated to leverage its embedded domain knowledge. Then, to establish an alignment between MolX and the LLM's textual input space, the whole model in which the LLM is frozen, is pre-trained with a versatile strategy including a diverse set of tasks. Extensive experimental evaluations demonstrate that our proposed method only introduces a small number of trainable parameters while outperforming baselines on various downstream molecule-related tasks ranging from molecule-to-text translation to retrosynthesis, with and without fine-tuning the LLM.
Are we making much progress? Revisiting chemical reaction yield prediction from an imbalanced regression perspective
Feb 06, 2024



Abstract:The yield of a chemical reaction quantifies the percentage of the target product formed in relation to the reactants consumed during the chemical reaction. Accurate yield prediction can guide chemists toward selecting high-yield reactions during synthesis planning, offering valuable insights before dedicating time and resources to wet lab experiments. While recent advancements in yield prediction have led to overall performance improvement across the entire yield range, an open challenge remains in enhancing predictions for high-yield reactions, which are of greater concern to chemists. In this paper, we argue that the performance gap in high-yield predictions results from the imbalanced distribution of real-world data skewed towards low-yield reactions, often due to unreacted starting materials and inherent ambiguities in the reaction processes. Despite this data imbalance, existing yield prediction methods continue to treat different yield ranges equally, assuming a balanced training distribution. Through extensive experiments on three real-world yield prediction datasets, we emphasize the urgent need to reframe reaction yield prediction as an imbalanced regression problem. Finally, we demonstrate that incorporating simple cost-sensitive re-weighting methods can significantly enhance the performance of yield prediction models on underrepresented high-yield regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge