Junjie Shan
Geminio: Language-Guided Gradient Inversion Attacks in Federated Learning
Nov 22, 2024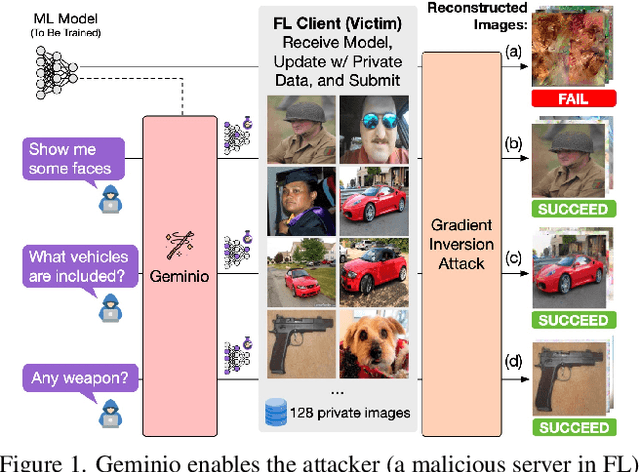

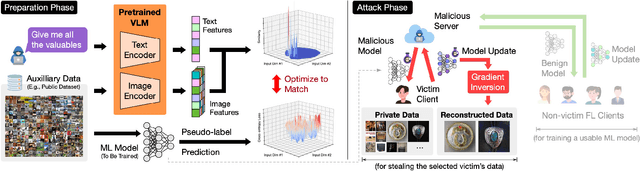
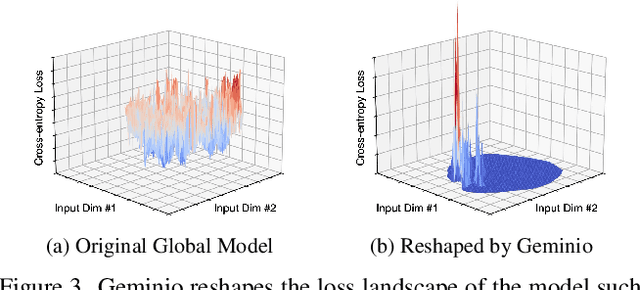
Abstract:Foundation models that bridge vision and language have made significant progress, inspiring numerous life-enriching applications. However, their potential for misuse to introduce new threats remains largely unexplored. This paper reveals that vision-language models (VLMs) can be exploited to overcome longstanding limitations in gradient inversion attacks (GIAs) within federated learning (FL), where an FL server reconstructs private data samples from gradients shared by victim clients. Current GIAs face challenges in reconstructing high-resolution images, especially when the victim has a large local data batch. While focusing reconstruction on valuable samples rather than the entire batch is promising, existing methods lack the flexibility to allow attackers to specify their target data. In this paper, we introduce Geminio, the first approach to transform GIAs into semantically meaningful, targeted attacks. Geminio enables a brand new privacy attack experience: attackers can describe, in natural language, the types of data they consider valuable, and Geminio will prioritize reconstruction to focus on those high-value samples. This is achieved by leveraging a pretrained VLM to guide the optimization of a malicious global model that, when shared with and optimized by a victim, retains only gradients of samples that match the attacker-specified query. Extensive experiments demonstrate Geminio's effectiveness in pinpointing and reconstructing targeted samples, with high success rates across complex datasets under FL and large batch sizes and showing resilience against existing defenses.
AnywhereDoor: Multi-Target Backdoor Attacks on Object Detection
Nov 21, 2024



Abstract:As object detection becomes integral to many safety-critical applications, understanding its vulnerabilities is essential. Backdoor attacks, in particular, pose a significant threat by implanting hidden backdoor in a victim model, which adversaries can later exploit to trigger malicious behaviors during inference. However, current backdoor techniques are limited to static scenarios where attackers must define a malicious objective before training, locking the attack into a predetermined action without inference-time adaptability. Given the expressive output space in object detection, including object existence detection, bounding box estimation, and object classification, the feasibility of implanting a backdoor that provides inference-time control with a high degree of freedom remains unexplored. This paper introduces AnywhereDoor, a flexible backdoor attack tailored for object detection. Once implanted, AnywhereDoor enables adversaries to specify different attack types (object vanishing, fabrication, or misclassification) and configurations (untargeted or targeted with specific classes) to dynamically control detection behavior. This flexibility is achieved through three key innovations: (i) objective disentanglement to support a broader range of attack combinations well beyond what existing methods allow; (ii) trigger mosaicking to ensure backdoor activations are robust, even against those object detectors that extract localized regions from the input image for recognition; and (iii) strategic batching to address object-level data imbalances that otherwise hinders a balanced manipulation. Extensive experiments demonstrate that AnywhereDoor provides attackers with a high degree of control, achieving an attack success rate improvement of nearly 80% compared to adaptations of existing methods for such flexible control.
BioKGBench: A Knowledge Graph Checking Benchmark of AI Agent for Biomedical Science
Jun 29, 2024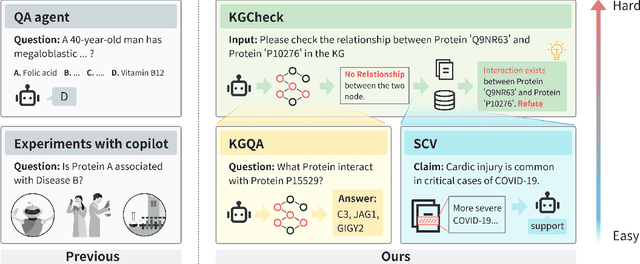
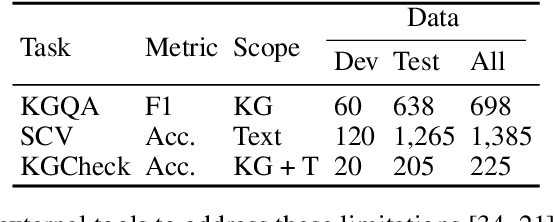
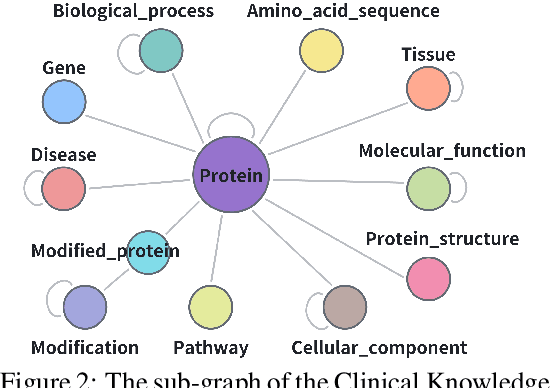
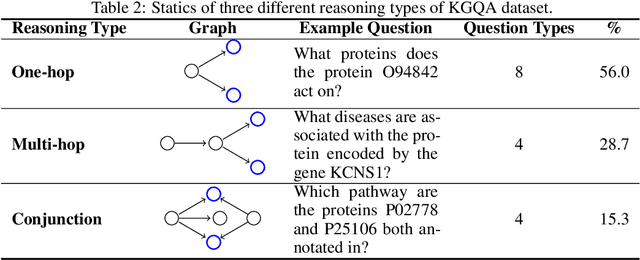
Abstract:Pursuing artificial intelligence for biomedical science, a.k.a. AI Scientist, draws increasing attention, where one common approach is to build a copilot agent driven by Large Language Models (LLMs). However, to evaluate such systems, people either rely on direct Question-Answering (QA) to the LLM itself, or in a biomedical experimental manner. How to precisely benchmark biomedical agents from an AI Scientist perspective remains largely unexplored. To this end, we draw inspiration from one most important abilities of scientists, understanding the literature, and introduce BioKGBench. In contrast to traditional evaluation benchmark that only focuses on factual QA, where the LLMs are known to have hallucination issues, we first disentangle "Understanding Literature" into two atomic abilities, i) "Understanding" the unstructured text from research papers by performing scientific claim verification, and ii) Ability to interact with structured Knowledge-Graph Question-Answering (KGQA) as a form of "Literature" grounding. We then formulate a novel agent task, dubbed KGCheck, using KGQA and domain-based Retrieval-Augmented Generation (RAG) to identify the factual errors of existing large-scale knowledge graph databases. We collect over two thousand data for two atomic tasks and 225 high-quality annotated data for the agent task. Surprisingly, we discover that state-of-the-art agents, both daily scenarios and biomedical ones, have either failed or inferior performance on our benchmark. We then introduce a simple yet effective baseline, dubbed BKGAgent. On the widely used popular knowledge graph, we discover over 90 factual errors which provide scenarios for agents to make discoveries and demonstrate the effectiveness of our approach. The code and data are available at https://github.com/westlake-autolab/BioKGBench.
StepCoder: Improve Code Generation with Reinforcement Learning from Compiler Feedback
Feb 05, 2024



Abstract:The advancement of large language models (LLMs) has significantly propelled the field of code generation. Previous work integrated reinforcement learning (RL) with compiler feedback for exploring the output space of LLMs to enhance code generation quality. However, the lengthy code generated by LLMs in response to complex human requirements makes RL exploration a challenge. Also, since the unit tests may not cover the complicated code, optimizing LLMs by using these unexecuted code snippets is ineffective. To tackle these challenges, we introduce StepCoder, a novel RL framework for code generation, consisting of two main components: CCCS addresses the exploration challenge by breaking the long sequences code generation task into a Curriculum of Code Completion Subtasks, while FGO only optimizes the model by masking the unexecuted code segments to provide Fine-Grained Optimization. In addition, we furthermore construct the APPS+ dataset for RL training, which is manually verified to ensure the correctness of unit tests. Experimental results show that our method improves the ability to explore the output space and outperforms state-of-the-art approaches in corresponding benchmarks. Our dataset APPS+ and StepCoder are available online.
CausalAPM: Generalizable Literal Disentanglement for NLU Debiasing
May 04, 2023



Abstract:Dataset bias, i.e., the over-reliance on dataset-specific literal heuristics, is getting increasing attention for its detrimental effect on the generalization ability of NLU models. Existing works focus on eliminating dataset bias by down-weighting problematic data in the training process, which induce the omission of valid feature information while mitigating bias. In this work, We analyze the causes of dataset bias from the perspective of causal inference and propose CausalAPM, a generalizable literal disentangling framework to ameliorate the bias problem from feature granularity. The proposed approach projects literal and semantic information into independent feature subspaces, and constrains the involvement of literal information in subsequent predictions. Extensive experiments on three NLP benchmarks (MNLI, FEVER, and QQP) demonstrate that our proposed framework significantly improves the OOD generalization performance while maintaining ID performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge