Xinna Lin
$\texttt{PatentAgent}$: Intelligent Agent for Automated Pharmaceutical Patent Analysis
Oct 25, 2024Abstract:Pharmaceutical patents play a vital role in biochemical industries, especially in drug discovery, providing researchers with unique early access to data, experimental results, and research insights. With the advancement of machine learning, patent analysis has evolved from manual labor to tasks assisted by automatic tools. However, there still lacks an unified agent that assists every aspect of patent analysis, from patent reading to core chemical identification. Leveraging the capabilities of Large Language Models (LLMs) to understand requests and follow instructions, we introduce the $\textbf{first}$ intelligent agent in this domain, $\texttt{PatentAgent}$, poised to advance and potentially revolutionize the landscape of pharmaceutical research. $\texttt{PatentAgent}$ comprises three key end-to-end modules -- $\textit{PA-QA}$, $\textit{PA-Img2Mol}$, and $\textit{PA-CoreId}$ -- that respectively perform (1) patent question-answering, (2) image-to-molecular-structure conversion, and (3) core chemical structure identification, addressing the essential needs of scientists and practitioners in pharmaceutical patent analysis. Each module of $\texttt{PatentAgent}$ demonstrates significant effectiveness with the updated algorithm and the synergistic design of $\texttt{PatentAgent}$ framework. $\textit{PA-Img2Mol}$ outperforms existing methods across CLEF, JPO, UOB, and USPTO patent benchmarks with an accuracy gain between 2.46% and 8.37% while $\textit{PA-CoreId}$ realizes accuracy improvement ranging from 7.15% to 7.62% on PatentNetML benchmark. Our code and dataset will be publicly available.
BioKGBench: A Knowledge Graph Checking Benchmark of AI Agent for Biomedical Science
Jun 29, 2024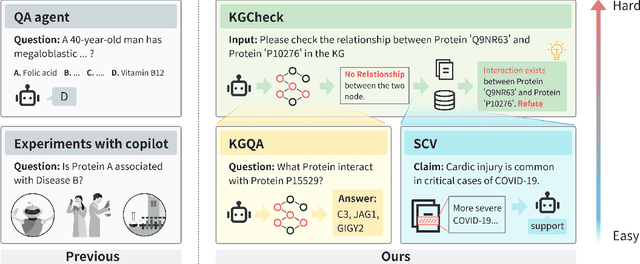
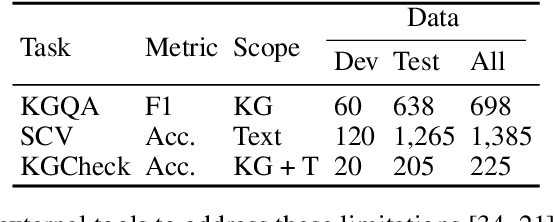
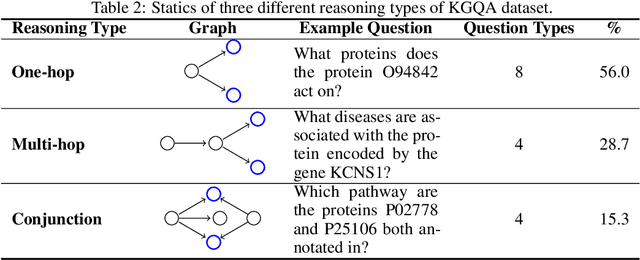
Abstract:Pursuing artificial intelligence for biomedical science, a.k.a. AI Scientist, draws increasing attention, where one common approach is to build a copilot agent driven by Large Language Models (LLMs). However, to evaluate such systems, people either rely on direct Question-Answering (QA) to the LLM itself, or in a biomedical experimental manner. How to precisely benchmark biomedical agents from an AI Scientist perspective remains largely unexplored. To this end, we draw inspiration from one most important abilities of scientists, understanding the literature, and introduce BioKGBench. In contrast to traditional evaluation benchmark that only focuses on factual QA, where the LLMs are known to have hallucination issues, we first disentangle "Understanding Literature" into two atomic abilities, i) "Understanding" the unstructured text from research papers by performing scientific claim verification, and ii) Ability to interact with structured Knowledge-Graph Question-Answering (KGQA) as a form of "Literature" grounding. We then formulate a novel agent task, dubbed KGCheck, using KGQA and domain-based Retrieval-Augmented Generation (RAG) to identify the factual errors of existing large-scale knowledge graph databases. We collect over two thousand data for two atomic tasks and 225 high-quality annotated data for the agent task. Surprisingly, we discover that state-of-the-art agents, both daily scenarios and biomedical ones, have either failed or inferior performance on our benchmark. We then introduce a simple yet effective baseline, dubbed BKGAgent. On the widely used popular knowledge graph, we discover over 90 factual errors which provide scenarios for agents to make discoveries and demonstrate the effectiveness of our approach. The code and data are available at https://github.com/westlake-autolab/BioKGBench.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge