Jiwon Lee
Comparison of Image Processing Models in Quark Gluon Jet Classification
Jan 29, 2026Abstract:We present a comprehensive comparison of convolutional and transformer-based models for distinguishing quark and gluon jets using simulated jet images from Pythia 8. By encoding jet substructure into a three-channel representation of particle kinematics, we evaluate the performance of convolutional neural networks (CNNs), Vision Transformers (ViTs), and Swin Transformers (Swin-Tiny) under both supervised and self-supervised learning setups. Our results show that fine-tuning only the final two transformer blocks of the Swin-Tiny model achieves the best trade-off between efficiency and accuracy, reaching 81.4% accuracy and an AUC (area under the ROC curve) of 88.9%. Self-supervised pretraining with Momentum Contrast (MoCo) further enhances feature robustness and reduces the number of trainable parameters. These findings highlight the potential of hierarchical attention-based models for jet substructure studies and for domain transfer to real collision data.
Unified Domain Generalization and Adaptation for Multi-View 3D Object Detection
Oct 29, 2024



Abstract:Recent advances in 3D object detection leveraging multi-view cameras have demonstrated their practical and economical value in various challenging vision tasks. However, typical supervised learning approaches face challenges in achieving satisfactory adaptation toward unseen and unlabeled target datasets (\ie, direct transfer) due to the inevitable geometric misalignment between the source and target domains. In practice, we also encounter constraints on resources for training models and collecting annotations for the successful deployment of 3D object detectors. In this paper, we propose Unified Domain Generalization and Adaptation (UDGA), a practical solution to mitigate those drawbacks. We first propose Multi-view Overlap Depth Constraint that leverages the strong association between multi-view, significantly alleviating geometric gaps due to perspective view changes. Then, we present a Label-Efficient Domain Adaptation approach to handle unfamiliar targets with significantly fewer amounts of labels (\ie, 1$\%$ and 5$\%)$, while preserving well-defined source knowledge for training efficiency. Overall, UDGA framework enables stable detection performance in both source and target domains, effectively bridging inevitable domain gaps, while demanding fewer annotations. We demonstrate the robustness of UDGA with large-scale benchmarks: nuScenes, Lyft, and Waymo, where our framework outperforms the current state-of-the-art methods.
A Learnable Counter-condition Analysis Framework for Functional Connectivity-based Neurological Disorder Diagnosis
Oct 06, 2023



Abstract:To understand the biological characteristics of neurological disorders with functional connectivity (FC), recent studies have widely utilized deep learning-based models to identify the disease and conducted post-hoc analyses via explainable models to discover disease-related biomarkers. Most existing frameworks consist of three stages, namely, feature selection, feature extraction for classification, and analysis, where each stage is implemented separately. However, if the results at each stage lack reliability, it can cause misdiagnosis and incorrect analysis in afterward stages. In this study, we propose a novel unified framework that systemically integrates diagnoses (i.e., feature selection and feature extraction) and explanations. Notably, we devised an adaptive attention network as a feature selection approach to identify individual-specific disease-related connections. We also propose a functional network relational encoder that summarizes the global topological properties of FC by learning the inter-network relations without pre-defined edges between functional networks. Last but not least, our framework provides a novel explanatory power for neuroscientific interpretation, also termed counter-condition analysis. We simulated the FC that reverses the diagnostic information (i.e., counter-condition FC): converting a normal brain to be abnormal and vice versa. We validated the effectiveness of our framework by using two large resting-state functional magnetic resonance imaging (fMRI) datasets, Autism Brain Imaging Data Exchange (ABIDE) and REST-meta-MDD, and demonstrated that our framework outperforms other competing methods for disease identification. Furthermore, we analyzed the disease-related neurological patterns based on counter-condition analysis.
GROW: A Row-Stationary Sparse-Dense GEMM Accelerator for Memory-Efficient Graph Convolutional Neural Networks
Mar 02, 2022



Abstract:Graph convolutional neural networks (GCNs) have emerged as a key technology in various application domains where the input data is relational. A unique property of GCNs is that its two primary execution stages, aggregation and combination, exhibit drastically different dataflows. Consequently, prior GCN accelerators tackle this research space by casting the aggregation and combination stages as a series of sparse-dense matrix multiplication. However, prior work frequently suffers from inefficient data movements, leaving significant performance left on the table. We present GROW, a GCN accelerator based on Gustavson's algorithm to architect a row-wise product based sparse-dense GEMM accelerator. GROW co-designs the software/hardware that strikes a balance in locality and parallelism for GCNs, achieving significant energy-efficiency improvements vs. state-of-the-art GCN accelerators.
Hybrid Graph Models for Logic Optimization via Spatio-Temporal Information
Jan 20, 2022



Abstract:Despite the stride made by machine learning (ML) based performance modeling, two major concerns that may impede production-ready ML applications in EDA are stringent accuracy requirements and generalization capability. To this end, we propose hybrid graph neural network (GNN) based approaches towards highly accurate quality-of-result (QoR) estimations with great generalization capability, specifically targeting logic synthesis optimization. The key idea is to simultaneously leverage spatio-temporal information from hardware designs and logic synthesis flows to forecast performance (i.e., delay/area) of various synthesis flows on different designs. The structural characteristics inside hardware designs are distilled and represented by GNNs; the temporal knowledge (i.e., relative ordering of logic transformations) in synthesis flows can be imposed on hardware designs by combining a virtually added supernode or a sequence processing model with conventional GNN models. Evaluation on 3.3 million data points shows that the testing mean absolute percentage error (MAPE) on designs seen and unseen during training are no more than 1.2% and 3.1%, respectively, which are 7-15X lower than existing studies.
hSDB-instrument: Instrument Localization Database for Laparoscopic and Robotic Surgeries
Oct 26, 2021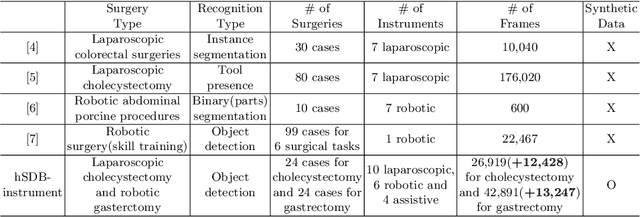
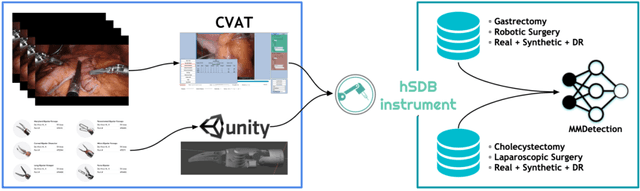
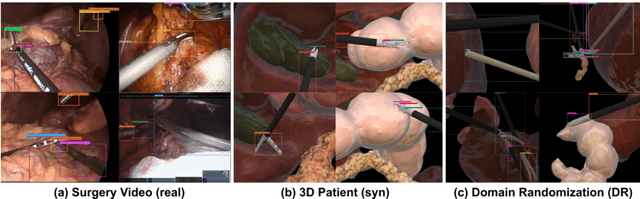
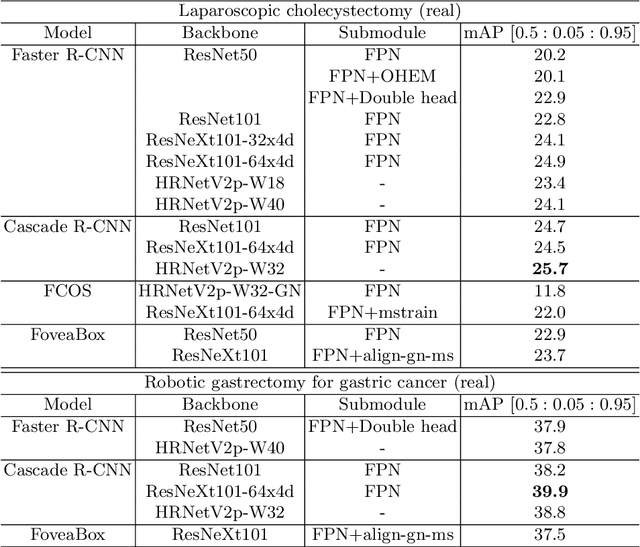
Abstract:Automated surgical instrument localization is an important technology to understand the surgical process and in order to analyze them to provide meaningful guidance during surgery or surgical index after surgery to the surgeon. We introduce a new dataset that reflects the kinematic characteristics of surgical instruments for automated surgical instrument localization of surgical videos. The hSDB(hutom Surgery DataBase)-instrument dataset consists of instrument localization information from 24 cases of laparoscopic cholecystecomy and 24 cases of robotic gastrectomy. Localization information for all instruments is provided in the form of a bounding box for object detection. To handle class imbalance problem between instruments, synthesized instruments modeled in Unity for 3D models are included as training data. Besides, for 3D instrument data, a polygon annotation is provided to enable instance segmentation of the tool. To reflect the kinematic characteristics of all instruments, they are annotated with head and body parts for laparoscopic instruments, and with head, wrist, and body parts for robotic instruments separately. Annotation data of assistive tools (specimen bag, needle, etc.) that are frequently used for surgery are also included. Moreover, we provide statistical information on the hSDB-instrument dataset and the baseline localization performances of the object detection networks trained by the MMDetection library and resulting analyses.
* https://hsdb-instrument.github.io
Rethinking Generalization Performance of Surgical Phase Recognition with Expert-Generated Annotations
Oct 22, 2021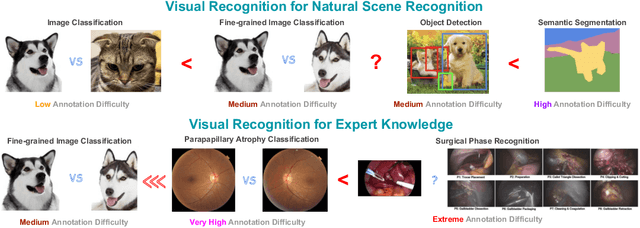
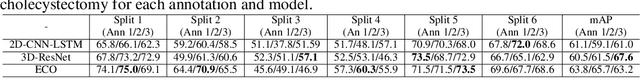

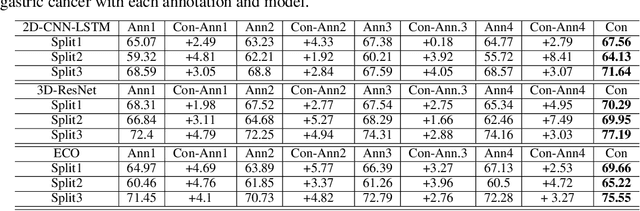
Abstract:As the area of application of deep neural networks expands to areas requiring expertise, e.g., in medicine and law, more exquisite annotation processes for expert knowledge training are required. In particular, it is difficult to guarantee generalization performance in the clinical field in the case of expert knowledge training where opinions may differ even among experts on annotations. To raise the issue of the annotation generation process for expertise training of CNNs, we verified the annotations for surgical phase recognition of laparoscopic cholecystectomy and subtotal gastrectomy for gastric cancer. We produce calibrated annotations for the seven phases of cholecystectomy by analyzing the discrepancies of previously annotated labels and by discussing the criteria of surgical phases. For gastrectomy for gastric cancer has more complex twenty-one surgical phases, we generate consensus annotation by the revision process with five specialists. By training the CNN-based surgical phase recognition networks with revised annotations, we achieved improved generalization performance over models trained with original annotation under the same cross-validation settings. We showed that the expertise data annotation pipeline for deep neural networks should be more rigorous based on the type of problem to apply clinical field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge