Jing Gong
HybridMamba: A Dual-domain Mamba for 3D Medical Image Segmentation
Sep 18, 2025Abstract:In the domain of 3D biomedical image segmentation, Mamba exhibits the superior performance for it addresses the limitations in modeling long-range dependencies inherent to CNNs and mitigates the abundant computational overhead associated with Transformer-based frameworks when processing high-resolution medical volumes. However, attaching undue importance to global context modeling may inadvertently compromise critical local structural information, thus leading to boundary ambiguity and regional distortion in segmentation outputs. Therefore, we propose the HybridMamba, an architecture employing dual complementary mechanisms: 1) a feature scanning strategy that progressively integrates representations both axial-traversal and local-adaptive pathways to harmonize the relationship between local and global representations, and 2) a gated module combining spatial-frequency analysis for comprehensive contextual modeling. Besides, we collect a multi-center CT dataset related to lung cancer. Experiments on MRI and CT datasets demonstrate that HybridMamba significantly outperforms the state-of-the-art methods in 3D medical image segmentation.
Training Compute-Optimal Protein Language Models
Nov 04, 2024Abstract:We explore optimally training protein language models, an area of significant interest in biological research where guidance on best practices is limited. Most models are trained with extensive compute resources until performance gains plateau, focusing primarily on increasing model sizes rather than optimizing the efficient compute frontier that balances performance and compute budgets. Our investigation is grounded in a massive dataset consisting of 939 million protein sequences. We trained over 300 models ranging from 3.5 million to 10.7 billion parameters on 5 to 200 billion unique tokens, to investigate the relations between model sizes, training token numbers, and objectives. First, we observed the effect of diminishing returns for the Causal Language Model (CLM) and that of overfitting for the Masked Language Model~(MLM) when repeating the commonly used Uniref database. To address this, we included metagenomic protein sequences in the training set to increase the diversity and avoid the plateau or overfitting effects. Second, we obtained the scaling laws of CLM and MLM on Transformer, tailored to the specific characteristics of protein sequence data. Third, we observe a transfer scaling phenomenon from CLM to MLM, further demonstrating the effectiveness of transfer through scaling behaviors based on estimated Effectively Transferred Tokens. Finally, to validate our scaling laws, we compare the large-scale versions of ESM-2 and PROGEN2 on downstream tasks, encompassing evaluations of protein generation as well as structure- and function-related tasks, all within less or equivalent pre-training compute budgets.
CoSQA+: Enhancing Code Search Dataset with Matching Code
Jun 17, 2024



Abstract:Semantic code search, retrieving code that matches a given natural language query, is an important task to improve productivity in software engineering. Existing code search datasets are problematic: either using unrealistic queries, or with mismatched codes, and typically using one-to-one query-code pairing, which fails to reflect the reality that a query might have multiple valid code matches. This paper introduces CoSQA+, pairing high-quality queries (reused from CoSQA) with multiple suitable codes. We collect code candidates from diverse sources and form candidate pairs by pairing queries with these codes. Utilizing the power of large language models (LLMs), we automate pair annotation, filtering, and code generation for queries without suitable matches. Through extensive experiments, CoSQA+ has demonstrated superior quality over CoSQA. Models trained on CoSQA+ exhibit improved performance. Furthermore, we propose a new metric Mean Multi-choice Reciprocal Rank (MMRR), to assess one-to-N code search performance. We provide the code and data at https://github.com/DeepSoftwareAnalytics/CoSQA_Plus.
xTrimoPGLM: Unified 100B-Scale Pre-trained Transformer for Deciphering the Language of Protein
Jan 11, 2024Abstract:Protein language models have shown remarkable success in learning biological information from protein sequences. However, most existing models are limited by either autoencoding or autoregressive pre-training objectives, which makes them struggle to handle protein understanding and generation tasks concurrently. We propose a unified protein language model, xTrimoPGLM, to address these two types of tasks simultaneously through an innovative pre-training framework. Our key technical contribution is an exploration of the compatibility and the potential for joint optimization of the two types of objectives, which has led to a strategy for training xTrimoPGLM at an unprecedented scale of 100 billion parameters and 1 trillion training tokens. Our extensive experiments reveal that 1) xTrimoPGLM significantly outperforms other advanced baselines in 18 protein understanding benchmarks across four categories. The model also facilitates an atomic-resolution view of protein structures, leading to an advanced 3D structural prediction model that surpasses existing language model-based tools. 2) xTrimoPGLM not only can generate de novo protein sequences following the principles of natural ones, but also can perform programmable generation after supervised fine-tuning (SFT) on curated sequences. These results highlight the substantial capability and versatility of xTrimoPGLM in understanding and generating protein sequences, contributing to the evolving landscape of foundation models in protein science.
xTrimoGene: An Efficient and Scalable Representation Learner for Single-Cell RNA-Seq Data
Nov 26, 2023Abstract:Advances in high-throughput sequencing technology have led to significant progress in measuring gene expressions at the single-cell level. The amount of publicly available single-cell RNA-seq (scRNA-seq) data is already surpassing 50M records for humans with each record measuring 20,000 genes. This highlights the need for unsupervised representation learning to fully ingest these data, yet classical transformer architectures are prohibitive to train on such data in terms of both computation and memory. To address this challenge, we propose a novel asymmetric encoder-decoder transformer for scRNA-seq data, called xTrimoGene$^\alpha$ (or xTrimoGene for short), which leverages the sparse characteristic of the data to scale up the pre-training. This scalable design of xTrimoGene reduces FLOPs by one to two orders of magnitude compared to classical transformers while maintaining high accuracy, enabling us to train the largest transformer models over the largest scRNA-seq dataset today. Our experiments also show that the performance of xTrimoGene improves as we scale up the model sizes, and it also leads to SOTA performance over various downstream tasks, such as cell type annotation, perturb-seq effect prediction, and drug combination prediction. xTrimoGene model is now available for use as a service via the following link: https://api.biomap.com/xTrimoGene/apply.
ApproxTrain: Fast Simulation of Approximate Multipliers for DNN Training and Inference
Sep 13, 2022



Abstract:Edge training of Deep Neural Networks (DNNs) is a desirable goal for continuous learning; however, it is hindered by the enormous computational power required by training. Hardware approximate multipliers have shown their effectiveness for gaining resource-efficiency in DNN inference accelerators; however, training with approximate multipliers is largely unexplored. To build resource efficient accelerators with approximate multipliers supporting DNN training, a thorough evaluation of training convergence and accuracy for different DNN architectures and different approximate multipliers is needed. This paper presents ApproxTrain, an open-source framework that allows fast evaluation of DNN training and inference using simulated approximate multipliers. ApproxTrain is as user-friendly as TensorFlow (TF) and requires only a high-level description of a DNN architecture along with C/C++ functional models of the approximate multiplier. We improve the speed of the simulation at the multiplier level by using a novel LUT-based approximate floating-point (FP) multiplier simulator on GPU (AMSim). ApproxTrain leverages CUDA and efficiently integrates AMSim into the TensorFlow library, in order to overcome the absence of native hardware approximate multiplier in commercial GPUs. We use ApproxTrain to evaluate the convergence and accuracy of DNN training with approximate multipliers for small and large datasets (including ImageNet) using LeNets and ResNets architectures. The evaluations demonstrate similar convergence behavior and negligible change in test accuracy compared to FP32 and bfloat16 multipliers. Compared to CPU-based approximate multiplier simulations in training and inference, the GPU-accelerated ApproxTrain is more than 2500x faster. Based on highly optimized closed-source cuDNN/cuBLAS libraries with native hardware multipliers, the original TensorFlow is only 8x faster than ApproxTrain.
Improving Machine Reading Comprehension with Single-choice Decision and Transfer Learning
Nov 06, 2020
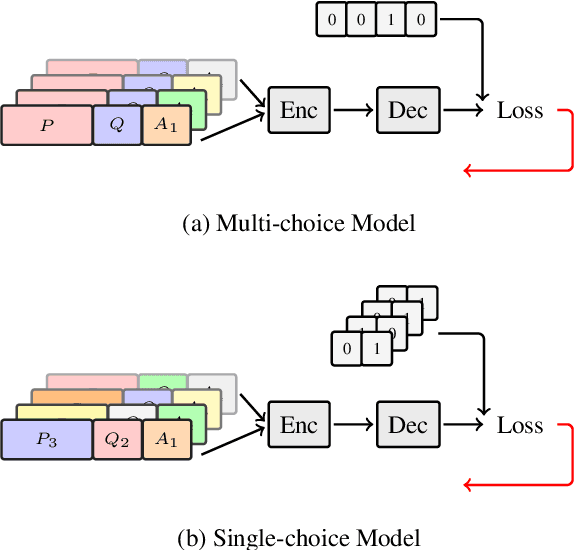
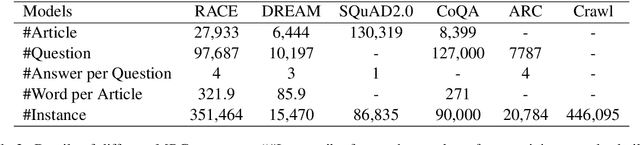
Abstract:Multi-choice Machine Reading Comprehension (MMRC) aims to select the correct answer from a set of options based on a given passage and question. Due to task specific of MMRC, it is no-trivial to transfer knowledge from other MRC tasks such as SQuAD, Dream. In this paper, we simply reconstruct multi-choice to single-choice by training a binary classification to distinguish whether a certain answer is correct. Then select the option with the highest confidence score. We construct our model upon ALBERT-xxlarge model and estimate it on the RACE dataset. During training, We adopt AutoML strategy to tune better parameters. Experimental results show that the single-choice is better than multi-choice. In addition, by transferring knowledge from other kinds of MRC tasks, our model achieves the new state of the art results in both single and ensemble settings.
Towards Scalable Distributed Training of Deep Learning on Public Cloud Clusters
Oct 20, 2020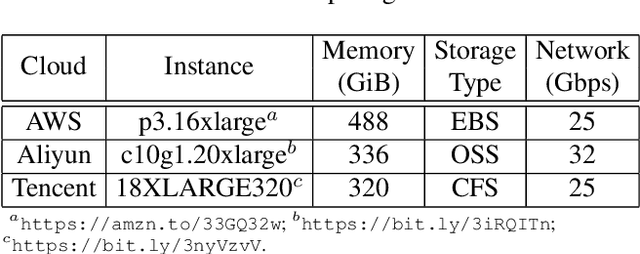
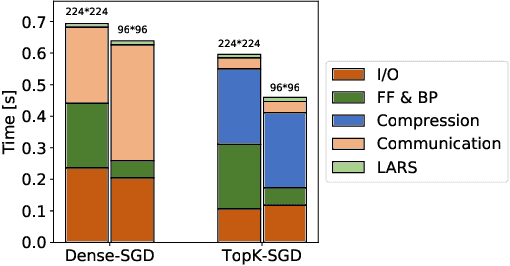

Abstract:Distributed training techniques have been widely deployed in large-scale deep neural networks (DNNs) training on dense-GPU clusters. However, on public cloud clusters, due to the moderate inter-connection bandwidth between instances, traditional state-of-the-art distributed training systems cannot scale well in training large-scale models. In this paper, we propose a new computing and communication efficient top-k sparsification communication library for distributed training. To further improve the system scalability, we optimize I/O by proposing a simple yet efficient multi-level data caching mechanism and optimize the update operation by introducing a novel parallel tensor operator. Experimental results on a 16-node Tencent Cloud cluster (each node with 8 Nvidia Tesla V100 GPUs) show that our system achieves 25%-40% faster than existing state-of-the-art systems on CNNs and Transformer. We finally break the record on DAWNBench on training ResNet-50 to 93% top-5 accuracy on ImageNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge