Jiaxi Wang
MIPS: a Multimodal Infinite Polymer Sequence Pre-training Framework for Polymer Property Prediction
Jul 27, 2025Abstract:Polymers, composed of repeating structural units called monomers, are fundamental materials in daily life and industry. Accurate property prediction for polymers is essential for their design, development, and application. However, existing modeling approaches, which typically represent polymers by the constituent monomers, struggle to capture the whole properties of polymer, since the properties change during the polymerization process. In this study, we propose a Multimodal Infinite Polymer Sequence (MIPS) pre-training framework, which represents polymers as infinite sequences of monomers and integrates both topological and spatial information for comprehensive modeling. From the topological perspective, we generalize message passing mechanism (MPM) and graph attention mechanism (GAM) to infinite polymer sequences. For MPM, we demonstrate that applying MPM to infinite polymer sequences is equivalent to applying MPM on the induced star-linking graph of monomers. For GAM, we propose to further replace global graph attention with localized graph attention (LGA). Moreover, we show the robustness of the "star linking" strategy through Repeat and Shift Invariance Test (RSIT). Despite its robustness, "star linking" strategy exhibits limitations when monomer side chains contain ring structures, a common characteristic of polymers, as it fails the Weisfeiler-Lehman~(WL) test. To overcome this issue, we propose backbone embedding to enhance the capability of MPM and LGA on infinite polymer sequences. From the spatial perspective, we extract 3D descriptors of repeating monomers to capture spatial information. Finally, we design a cross-modal fusion mechanism to unify the topological and spatial information. Experimental validation across eight diverse polymer property prediction tasks reveals that MIPS achieves state-of-the-art performance.
Enhancing Multi-task Learning Capability of Medical Generalist Foundation Model via Image-centric Multi-annotation Data
Apr 14, 2025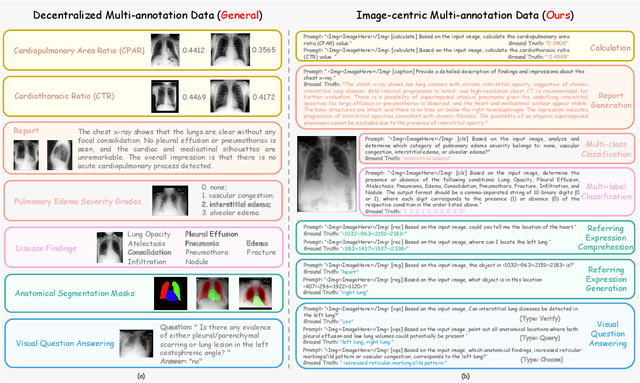
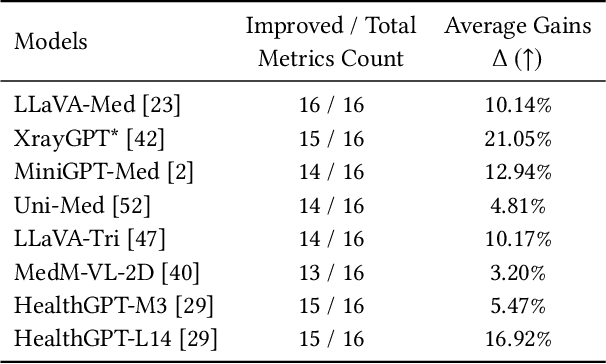
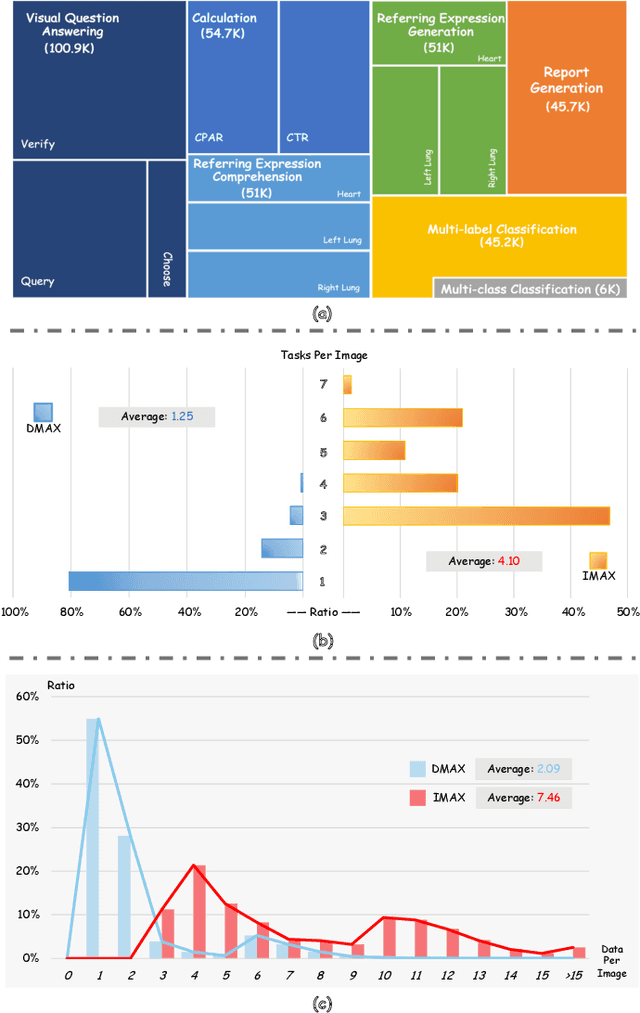
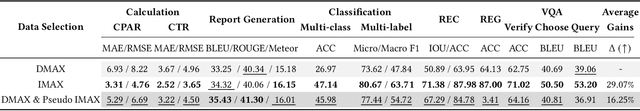
Abstract:The emergence of medical generalist foundation models has revolutionized conventional task-specific model development paradigms, aiming to better handle multiple tasks through joint training on large-scale medical datasets. However, recent advances prioritize simple data scaling or architectural component enhancement, while neglecting to re-examine multi-task learning from a data-centric perspective. Critically, simply aggregating existing data resources leads to decentralized image-task alignment, which fails to cultivate comprehensive image understanding or align with clinical needs for multi-dimensional image interpretation. In this paper, we introduce the image-centric multi-annotation X-ray dataset (IMAX), the first attempt to enhance the multi-task learning capabilities of medical multi-modal large language models (MLLMs) from the data construction level. To be specific, IMAX is featured from the following attributes: 1) High-quality data curation. A comprehensive collection of more than 354K entries applicable to seven different medical tasks. 2) Image-centric dense annotation. Each X-ray image is associated with an average of 4.10 tasks and 7.46 training entries, ensuring multi-task representation richness per image. Compared to the general decentralized multi-annotation X-ray dataset (DMAX), IMAX consistently demonstrates significant multi-task average performance gains ranging from 3.20% to 21.05% across seven open-source state-of-the-art medical MLLMs. Moreover, we investigate differences in statistical patterns exhibited by IMAX and DMAX training processes, exploring potential correlations between optimization dynamics and multi-task performance. Finally, leveraging the core concept of IMAX data construction, we propose an optimized DMAX-based training strategy to alleviate the dilemma of obtaining high-quality IMAX data in practical scenarios.
Bridging Modality Gap for Visual Grounding with Effecitve Cross-modal Distillation
Dec 29, 2023Abstract:Visual grounding aims to align visual information of specific regions of images with corresponding natural language expressions. Current visual grounding methods leverage pre-trained visual and language backbones separately to obtain visual features and linguistic features. Although these two types of features are then fused via delicately designed networks, the heterogeneity of the features makes them inapplicable for multi-modal reasoning. This problem arises from the domain gap between the single-modal pre-training backbone used in current visual grounding methods, which can hardly be overcome by the traditional end-to-end training method. To alleviate this, our work proposes an Empowering pre-trained model for Visual Grounding (EpmVG) framework, which distills a multimodal pre-trained model to guide the visual grounding task. EpmVG is based on a novel cross-modal distillation mechanism, which can effectively introduce the consistency information of images and texts in the pre-trained model, to reduce the domain gap existing in the backbone networks, thereby improving the performance of the model in the visual grounding task. Extensive experiments are carried out on five conventionally used datasets, and results demonstrate that our method achieves better performance than state-of-the-art methods.
Towards Predicting Equilibrium Distributions for Molecular Systems with Deep Learning
Jun 08, 2023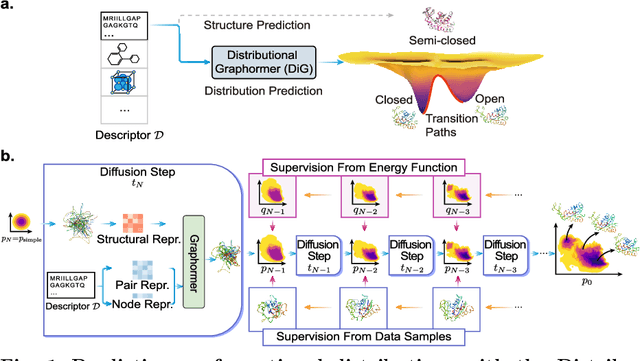
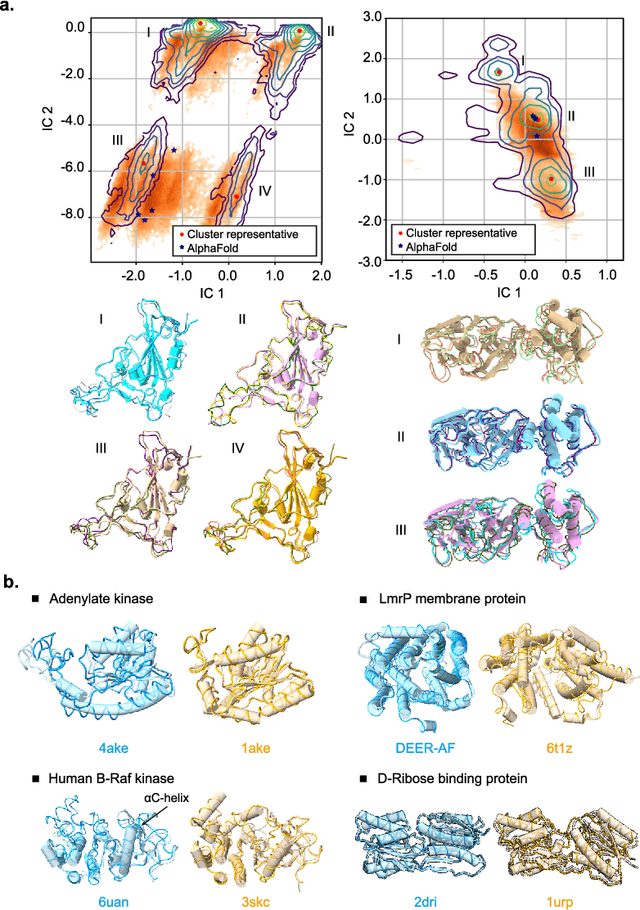
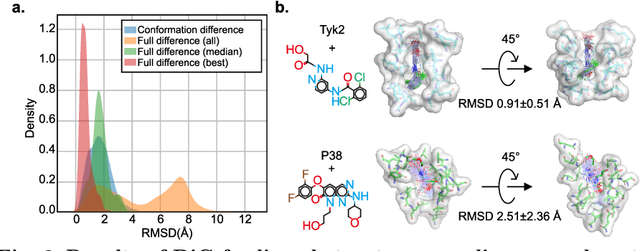
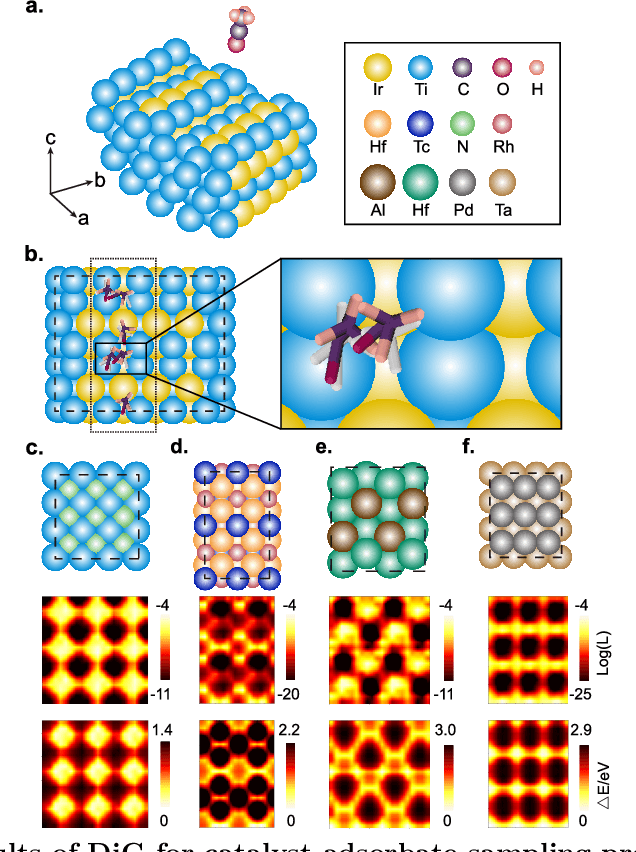
Abstract:Advances in deep learning have greatly improved structure prediction of molecules. However, many macroscopic observations that are important for real-world applications are not functions of a single molecular structure, but rather determined from the equilibrium distribution of structures. Traditional methods for obtaining these distributions, such as molecular dynamics simulation, are computationally expensive and often intractable. In this paper, we introduce a novel deep learning framework, called Distributional Graphormer (DiG), in an attempt to predict the equilibrium distribution of molecular systems. Inspired by the annealing process in thermodynamics, DiG employs deep neural networks to transform a simple distribution towards the equilibrium distribution, conditioned on a descriptor of a molecular system, such as a chemical graph or a protein sequence. This framework enables efficient generation of diverse conformations and provides estimations of state densities. We demonstrate the performance of DiG on several molecular tasks, including protein conformation sampling, ligand structure sampling, catalyst-adsorbate sampling, and property-guided structure generation. DiG presents a significant advancement in methodology for statistically understanding molecular systems, opening up new research opportunities in molecular science.
Understanding the Failure of Batch Normalization for Transformers in NLP
Oct 11, 2022



Abstract:Batch Normalization (BN) is a core and prevalent technique in accelerating the training of deep neural networks and improving the generalization on Computer Vision (CV) tasks. However, it fails to defend its position in Natural Language Processing (NLP), which is dominated by Layer Normalization (LN). In this paper, we are trying to answer why BN usually performs worse than LN in NLP tasks with Transformer models. We find that the inconsistency between training and inference of BN is the leading cause that results in the failure of BN in NLP. We define Training Inference Discrepancy (TID) to quantitatively measure this inconsistency and reveal that TID can indicate BN's performance, supported by extensive experiments, including image classification, neural machine translation, language modeling, sequence labeling, and text classification tasks. We find that BN can obtain much better test performance than LN when TID keeps small through training. To suppress the explosion of TID, we propose Regularized BN (RBN) that adds a simple regularization term to narrow the gap between batch statistics and population statistics of BN. RBN improves the performance of BN consistently and outperforms or is on par with LN on 17 out of 20 settings, involving ten datasets and two common variants of Transformer\footnote{Our code is available at \url{https://github.com/wjxts/RegularizedBN}}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge