Jianyu Lin
SIGMA: Scalable Spectral Insights for LLM Collapse
Jan 06, 2026Abstract:The rapid adoption of synthetic data for training Large Language Models (LLMs) has introduced the technical challenge of "model collapse"-a degenerative process where recursive training on model-generated content leads to a contraction of distributional variance and representational quality. While the phenomenology of collapse is increasingly evident, rigorous methods to quantify and predict its onset in high-dimensional spaces remain elusive. In this paper, we introduce SIGMA (Spectral Inequalities for Gram Matrix Analysis), a unified framework that benchmarks model collapse through the spectral lens of the embedding Gram matrix. By deriving and utilizing deterministic and stochastic bounds on the matrix's spectrum, SIGMA provides a mathematically grounded metric to track the contraction of the representation space. Crucially, our stochastic formulation enables scalable estimation of these bounds, making the framework applicable to large-scale foundation models where full eigendecomposition is intractable. We demonstrate that SIGMA effectively captures the transition towards degenerate states, offering both theoretical insights into the mechanics of collapse and a practical, scalable tool for monitoring the health of recursive training pipelines.
BDIS-SLAM: A lightweight CPU-based dense stereo SLAM for surgery
Dec 25, 2023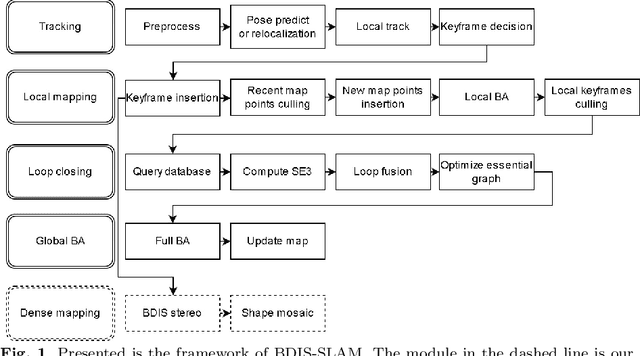
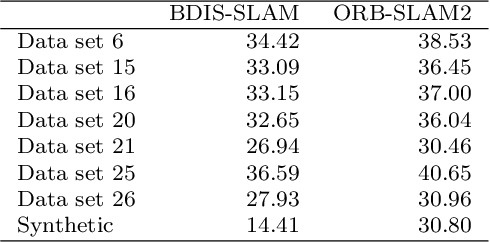
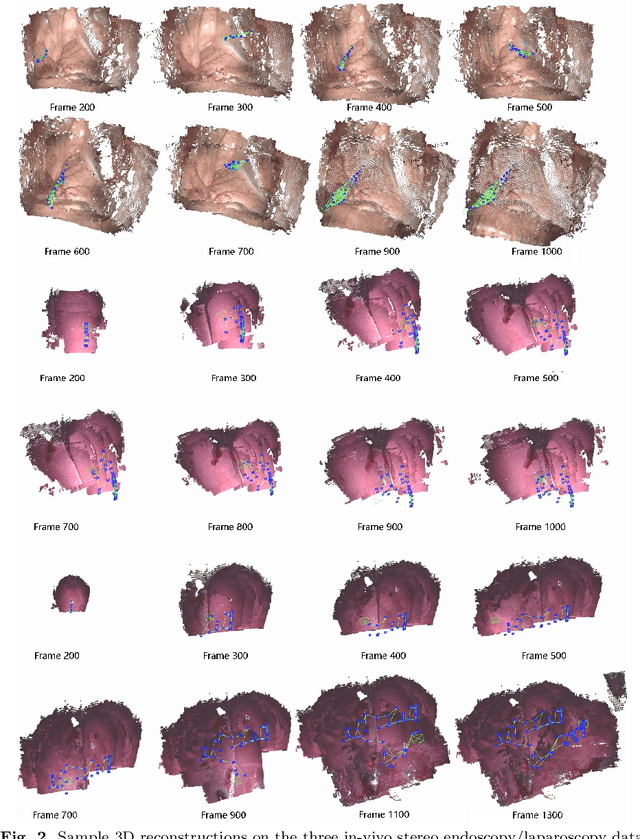
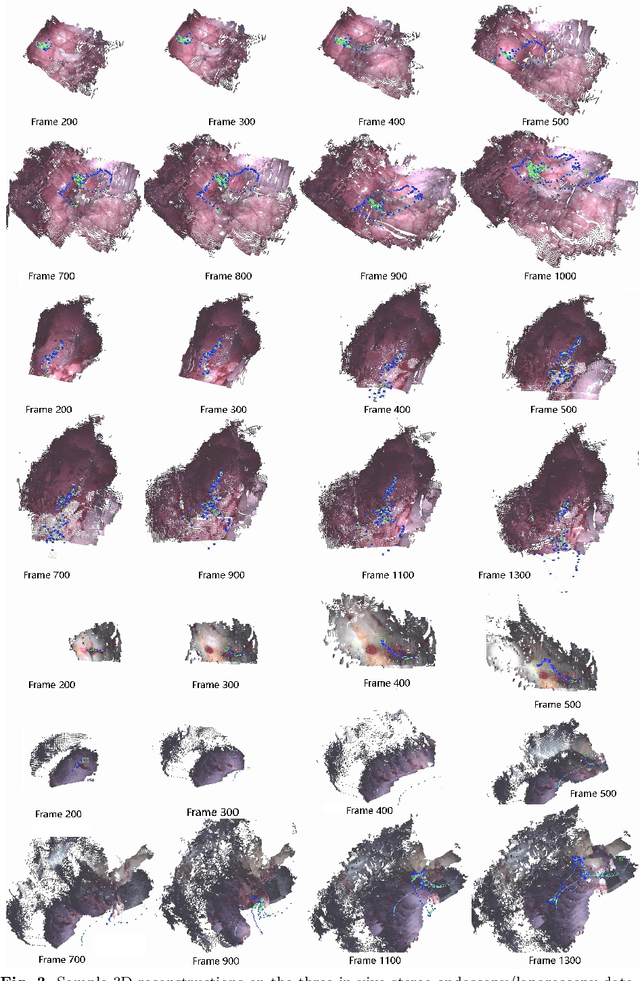
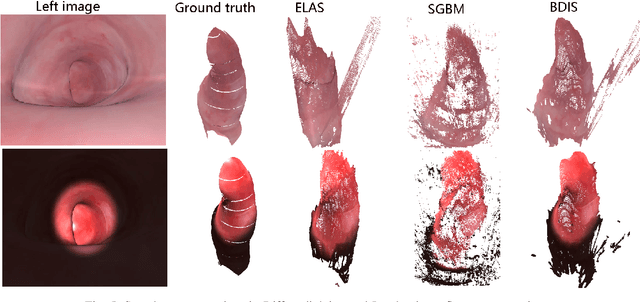
Abstract:Purpose: Common dense stereo Simultaneous Localization and Mapping (SLAM) approaches in Minimally Invasive Surgery (MIS) require high-end parallel computational resources for real-time implementation. Yet, it is not always feasible since the computational resources should be allocated to other tasks like segmentation, detection, and tracking. To solve the problem of limited parallel computational power, this research aims at a lightweight dense stereo SLAM system that works on a single-core CPU and achieves real-time performance (more than 30 Hz in typical scenarios). Methods: A new dense stereo mapping module is integrated with the ORB-SLAM2 system and named BDIS-SLAM. Our new dense stereo mapping module includes stereo matching and 3D dense depth mosaic methods. Stereo matching is achieved with the recently proposed CPU-level real-time matching algorithm Bayesian Dense Inverse Searching (BDIS). A BDIS-based shape recovery and a depth mosaic strategy are integrated as a new thread and coupled with the backbone ORB-SLAM2 system for real-time stereo shape recovery. Results: Experiments on in-vivo data sets show that BDIS-SLAM runs at over 30 Hz speed on modern single-core CPU in typical endoscopy/colonoscopy scenarios. BDIS-SLAM only consumes around an additional 12% time compared with the backbone ORB-SLAM2. Although our lightweight BDIS-SLAM simplifies the process by ignoring deformation and fusion procedures, it can provide a usable dense mapping for modern MIS on computationally constrained devices. Conclusion: The proposed BDIS-SLAM is a lightweight stereo dense SLAM system for MIS. It achieves 30 Hz on a modern single-core CPU in typical endoscopy/colonoscopy scenarios (image size around 640*480). BDIS-SLAM provides a low-cost solution for dense mapping in MIS and has the potential to be applied in surgical robots and AR systems.
BDIS: Bayesian Dense Inverse Searching Method for Real-Time Stereo Surgical Image Matching
May 06, 2022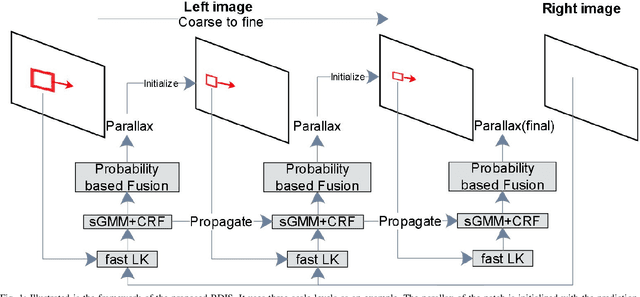

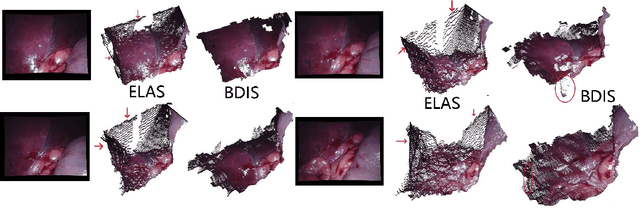
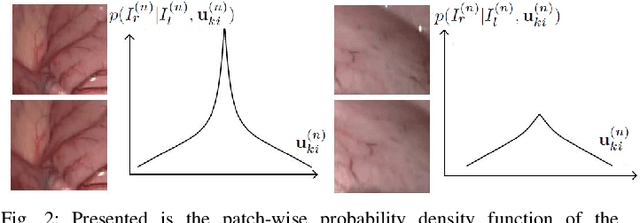
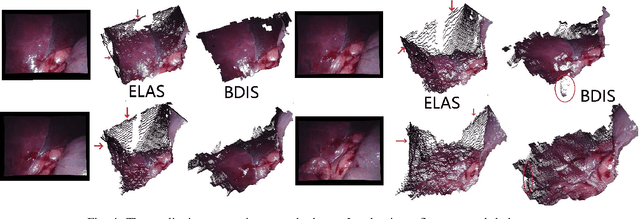
Abstract:In stereoscope-based Minimally Invasive Surgeries (MIS), dense stereo matching plays an indispensable role in 3D shape recovery, AR, VR, and navigation tasks. Although numerous Deep Neural Network (DNN) approaches are proposed, the conventional prior-free approaches are still popular in the industry because of the lack of open-source annotated data set and the limitation of the task-specific pre-trained DNNs. Among the prior-free stereo matching algorithms, there is no successful real-time algorithm in none GPU environment for MIS. This paper proposes the first CPU-level real-time prior-free stereo matching algorithm for general MIS tasks. We achieve an average 17 Hz on 640*480 images with a single-core CPU (i5-9400) for surgical images. Meanwhile, it achieves slightly better accuracy than the popular ELAS. The patch-based fast disparity searching algorithm is adopted for the rectified stereo images. A coarse-to-fine Bayesian probability and a spatial Gaussian mixed model were proposed to evaluate the patch probability at different scales. An optional probability density function estimation algorithm was adopted to quantify the prediction variance. Extensive experiments demonstrated the proposed method's capability to handle ambiguities introduced by the textureless surfaces and the photometric inconsistency from the non-Lambertian reflectance and dark illumination. The estimated probability managed to balance the confidences of the patches for stereo images at different scales. It has similar or higher accuracy and fewer outliers than the baseline ELAS in MIS, while it is 4-5 times faster. The code and the synthetic data sets are available at https://github.com/JingweiSong/BDIS-v2.
Bayesian dense inverse searching algorithm for real-time stereo matching in minimally invasive surgery
Jun 14, 2021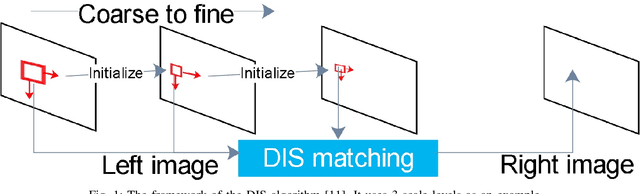
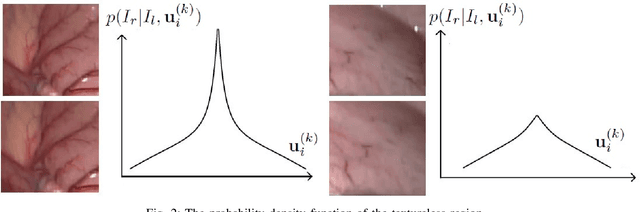
Abstract:This paper reports a CPU-level real-time stereo matching method for surgical images (10 Hz on 640 * 480 image with a single core of i5-9400). The proposed method is built on the fast ''dense inverse searching'' algorithm, which estimates the disparity of the stereo images. The overlapping image patches (arbitrary squared image segment) from the images at different scales are aligned based on the photometric consistency presumption. We propose a Bayesian framework to evaluate the probability of the optimized patch disparity at different scales. Moreover, we introduce a spatial Gaussian mixed probability distribution to address the pixel-wise probability within the patch. In-vivo and synthetic experiments show that our method can handle ambiguities resulted from the textureless surfaces and the photometric inconsistency caused by the Lambertian reflectance. Our Bayesian method correctly balances the probability of the patch for stereo images at different scales. Experiments indicate that the estimated depth has higher accuracy and fewer outliers than the baseline methods in the surgical scenario.
Estimation of Tissue Oxygen Saturation from RGB images and Sparse Hyperspectral Signals based on Conditional Generative Adversarial Network
May 16, 2019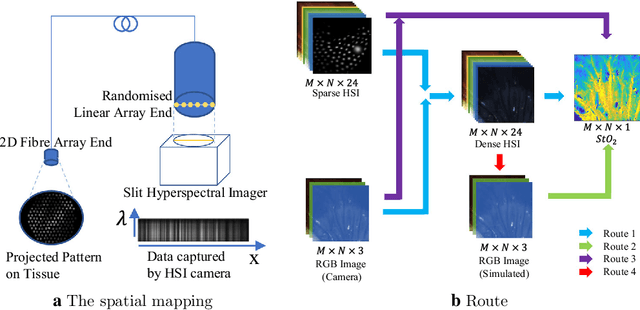
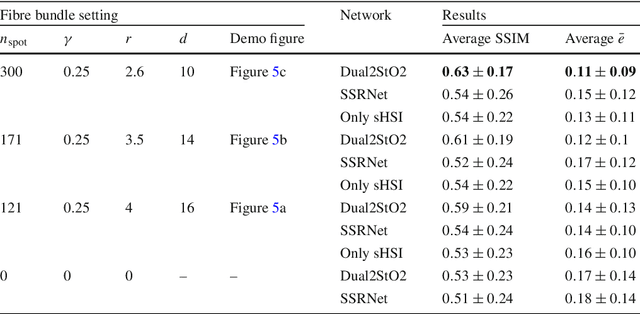

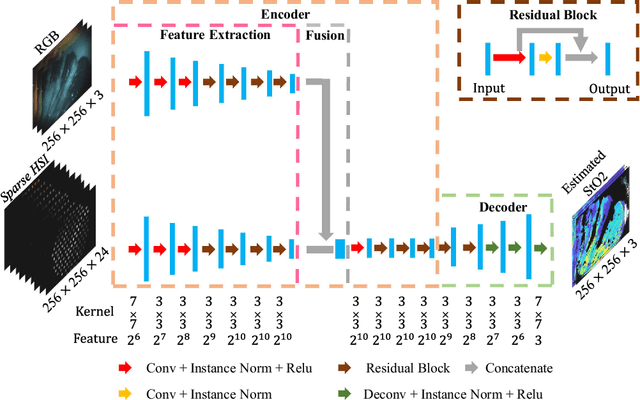
Abstract:Purpose: Intra-operative measurement of tissue oxygen saturation (StO2) is important in the detection of ischemia, monitoring perfusion and identifying disease. Hyperspectral imaging (HSI) measures the optical reflectance spectrum of the tissue and uses this information to quantify its composition, including StO2. However, real-time monitoring is difficult due to the capture rate and data processing time. Methods: An endoscopic system based on a multi-fiber probe was previously developed to sparsely capture HSI data (sHSI). These were combined with RGB images, via a deep neural network, to generate high-resolution hypercubes and calculate StO2. To improve accuracy and processing speed, we propose a dual-input conditional generative adversarial network (cGAN), Dual2StO2, to directly estimate StO2 by fusing features from both RGB and sHSI. Results: Validation experiments were carried out on in vivo porcine bowel data, where the ground truth StO2 was generated from the HSI camera. The performance was also compared to our previous super-spectral-resolution network, SSRNet in terms of mean StO2 prediction accuracy and structural similarity metrics. Dual2StO2 was also tested using simulated probe data with varying fiber number. Conclusions: StO2 estimation by Dual2StO2 is visually closer to ground truth in general structure, achieves higher prediction accuracy and faster processing speed than SSRNet. Simulations showed that results improved when a greater number of fibers are used in the probe. Future work will include refinement of the network architecture, hardware optimization based on simulation results, and evaluation of the technique in clinical applications beyond StO2 estimation.
Estimation of Tissue Oxygen Saturation from RGB Images based on Pixel-level Image Translation
Apr 19, 2018

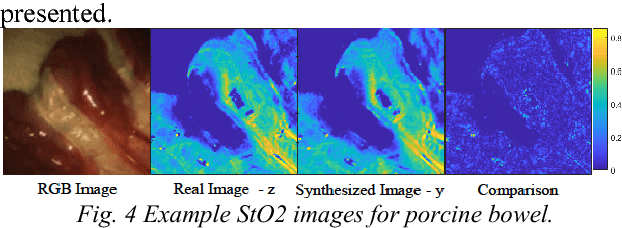
Abstract:Intra-operative measurement of tissue oxygen saturation (StO2) has been widely explored by pulse oximetry or hyperspectral imaging (HSI) to assess the function and viability of tissue. In this paper we propose a pixel- level image-to-image translation approach based on conditional Generative Adversarial Networks (cGAN) to estimate tissue oxygen saturation (StO2) directly from RGB images. The real-time performance and non-reliance on additional hardware, enable a seamless integration of the proposed method into surgical and diagnostic workflows with standard endoscope systems. For validation, RGB images and StO2 ground truth were simulated and estimated from HSI images collected by a liquid crystal tuneable filter (LCTF) endoscope for three tissue types (porcine bowel, lamb uterus and rabbit uterus). The result show that the proposed method can achieve visually identical images with comparable accuracy.
Real-time 3D Shape Instantiation from Single Fluoroscopy Projection for Fenestrated Stent Graft Deployment
Jan 08, 2018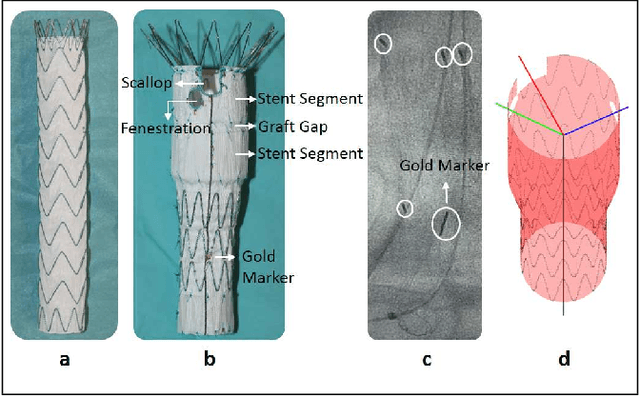
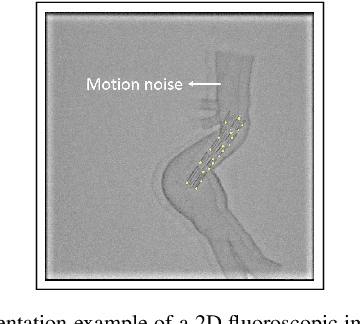
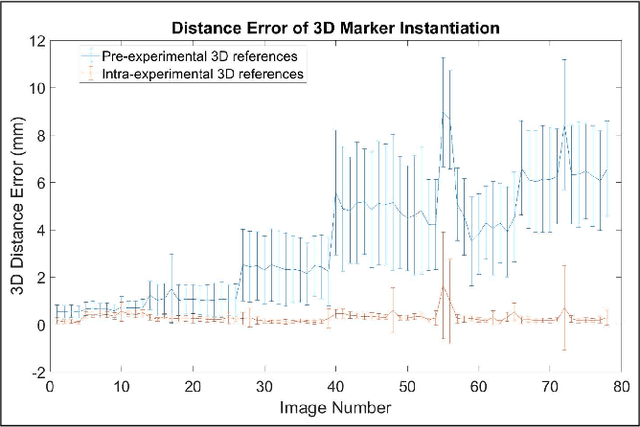
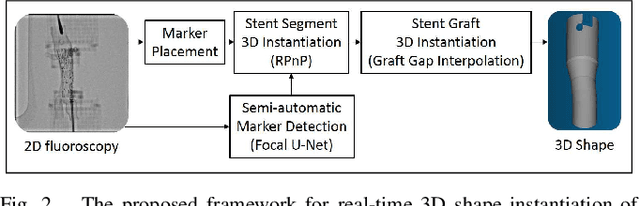
Abstract:Robot-assisted deployment of fenestrated stent grafts in Fenestrated Endovascular Aortic Repair (FEVAR) requires accurate geometrical alignment. Currently, this process is guided by 2D fluoroscopy, which is uninformative and error prone. In this paper, a real-time framework is proposed to instantiate the 3D shape of a fenestrated stent graft based on only a single low-dose 2D fluoroscopic image. Firstly, the fenestrated stent graft was placed with markers. Secondly, the 3D pose of each stent segment was instantiated by the RPnP (Robust Perspective-n-Point) method. Thirdly, the 3D shape of the whole stent graft was instantiated via graft gap interpolation. Focal-Unet was proposed to segment the markers from 2D fluoroscopic images to achieve semi-automatic marker detection. The proposed framework was validated on five patient-specific 3D printed phantoms of aortic aneurysms and three stent grafts with new marker placements, showing an average distance error of 1-3mm and an average angle error of 4 degree.
* 7 pages, 10 figures
Endoscopic Depth Measurement and Super-Spectral-Resolution Imaging
Jun 21, 2017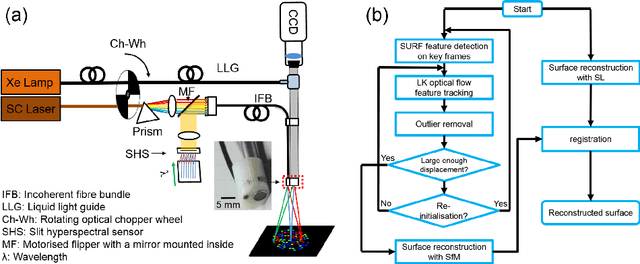

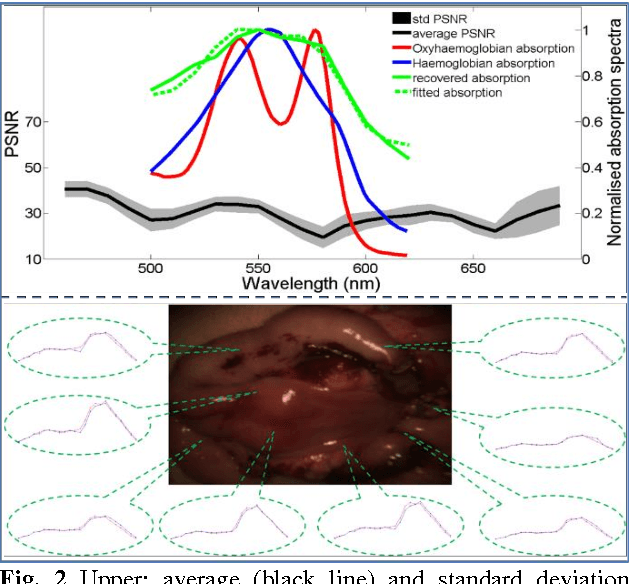
Abstract:Intra-operative measurements of tissue shape and multi/ hyperspectral information have the potential to provide surgical guidance and decision making support. We report an optical probe based system to combine sparse hyperspectral measurements and spectrally-encoded structured lighting (SL) for surface measurements. The system provides informative signals for navigation with a surgical interface. By rapidly switching between SL and white light (WL) modes, SL information is combined with structure-from-motion (SfM) from white light images, based on SURF feature detection and Lucas-Kanade (LK) optical flow to provide quasi-dense surface shape reconstruction with known scale in real-time. Furthermore, "super-spectral-resolution" was realized, whereby the RGB images and sparse hyperspectral data were integrated to recover dense pixel-level hyperspectral stacks, by using convolutional neural networks to upscale the wavelength dimension. Validation and demonstration of this system is reported on ex vivo/in vivo animal/ human experiments.
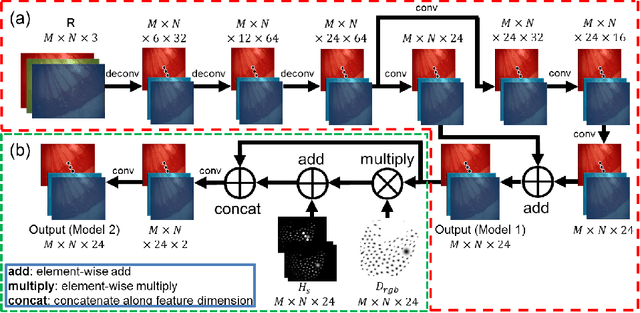
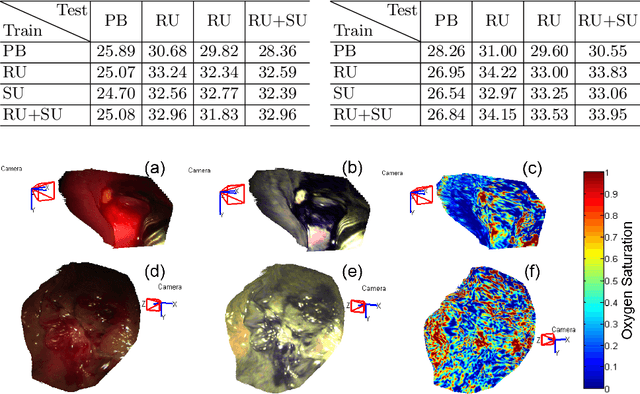
Recovering Dense Tissue Multispectral Signal from in vivo RGB Images
Jun 20, 2017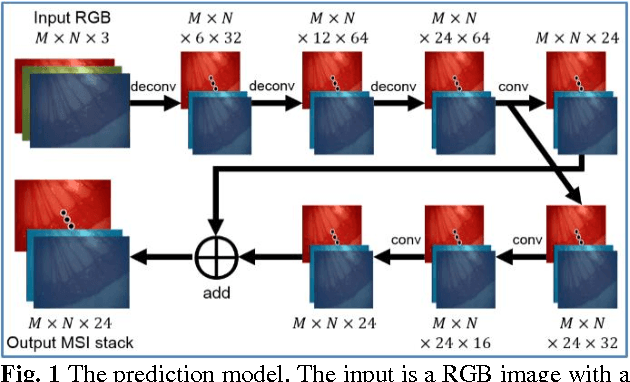
Abstract:Hyperspectral/multispectral imaging (HSI/MSI) contains rich information clinical applications, such as 1) narrow band imaging for vascular visualisation; 2) oxygen saturation for intraoperative perfusion monitoring and clinical decision making [1]; 3) tissue classification and identification of pathology [2]. The current systems which provide pixel-level HSI/MSI signal can be generally divided into two types: spatial scanning and spectral scanning. However, the trade-off between spatial/spectral resolution, the acquisition time, and the hardware complexity hampers implementation in real-world applications, especially intra-operatively. Acquiring high resolution images in real-time is important for HSI/MSI in intra-operative imaging, to alleviate the side effect caused by breathing, heartbeat, and other sources of motion. Therefore, we developed an algorithm to recover a pixel-level MSI stack using only the captured snapshot RGB images from a normal camera. We refer to this technique as "super-spectral-resolution". The proposed method enables recovery of pixel-level-dense MSI signals with 24 spectral bands at ~11 frames per second (FPS) on a GPU. Multispectral data captured from porcine bowel and sheep/rabbit uteri in vivo has been used for training, and the algorithm has been validated using unseen in vivo animal experiments.
Probe-based Rapid Hybrid Hyperspectral and Tissue Surface Imaging Aided by Fully Convolutional Networks
Jun 15, 2016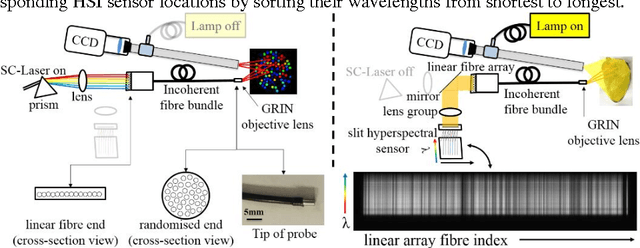
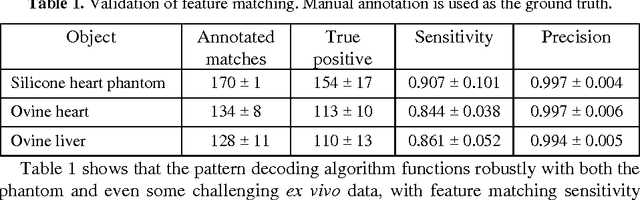
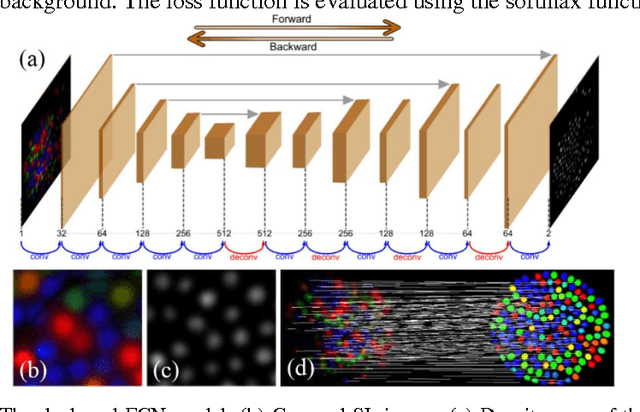
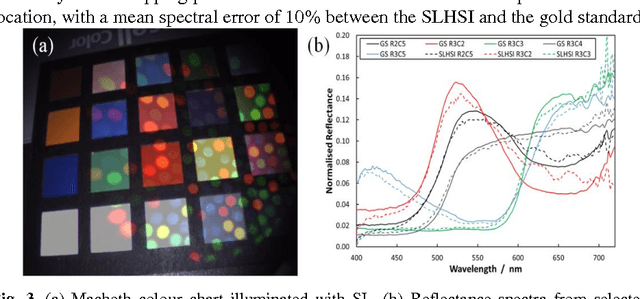
Abstract:Tissue surface shape and reflectance spectra provide rich intra-operative information useful in surgical guidance. We propose a hybrid system which displays an endoscopic image with a fast joint inspection of tissue surface shape using structured light (SL) and hyperspectral imaging (HSI). For SL a miniature fibre probe is used to project a coloured spot pattern onto the tissue surface. In HSI mode standard endoscopic illumination is used, with the fibre probe collecting reflected light and encoding the spatial information into a linear format that can be imaged onto the slit of a spectrograph. Correspondence between the arrangement of fibres at the distal and proximal ends of the bundle was found using spectral encoding. Then during pattern decoding, a fully convolutional network (FCN) was used for spot detection, followed by a matching propagation algorithm for spot identification. This method enabled fast reconstruction (12 frames per second) using a GPU. The hyperspectral image was combined with the white light image and the reconstructed surface, showing the spectral information of different areas. Validation of this system using phantom and ex vivo experiments has been demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge