Neil T. Clancy
Estimation of Tissue Oxygen Saturation from RGB images and Sparse Hyperspectral Signals based on Conditional Generative Adversarial Network
May 16, 2019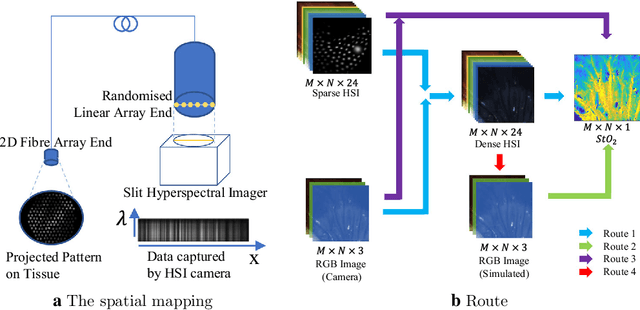
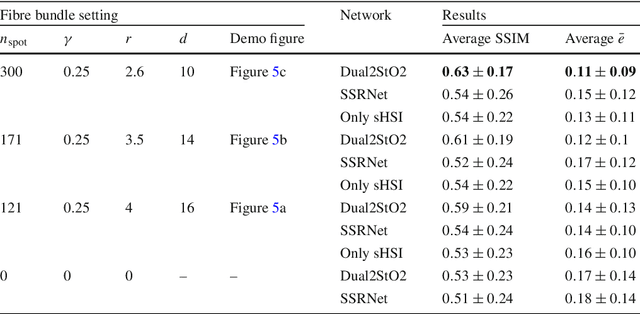

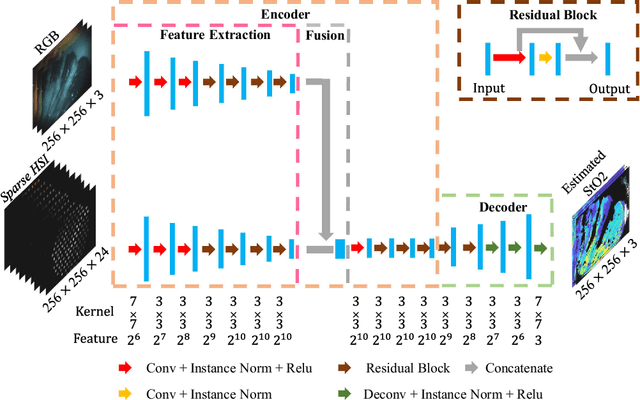
Abstract:Purpose: Intra-operative measurement of tissue oxygen saturation (StO2) is important in the detection of ischemia, monitoring perfusion and identifying disease. Hyperspectral imaging (HSI) measures the optical reflectance spectrum of the tissue and uses this information to quantify its composition, including StO2. However, real-time monitoring is difficult due to the capture rate and data processing time. Methods: An endoscopic system based on a multi-fiber probe was previously developed to sparsely capture HSI data (sHSI). These were combined with RGB images, via a deep neural network, to generate high-resolution hypercubes and calculate StO2. To improve accuracy and processing speed, we propose a dual-input conditional generative adversarial network (cGAN), Dual2StO2, to directly estimate StO2 by fusing features from both RGB and sHSI. Results: Validation experiments were carried out on in vivo porcine bowel data, where the ground truth StO2 was generated from the HSI camera. The performance was also compared to our previous super-spectral-resolution network, SSRNet in terms of mean StO2 prediction accuracy and structural similarity metrics. Dual2StO2 was also tested using simulated probe data with varying fiber number. Conclusions: StO2 estimation by Dual2StO2 is visually closer to ground truth in general structure, achieves higher prediction accuracy and faster processing speed than SSRNet. Simulations showed that results improved when a greater number of fibers are used in the probe. Future work will include refinement of the network architecture, hardware optimization based on simulation results, and evaluation of the technique in clinical applications beyond StO2 estimation.
Estimation of Tissue Oxygen Saturation from RGB Images based on Pixel-level Image Translation
Apr 19, 2018

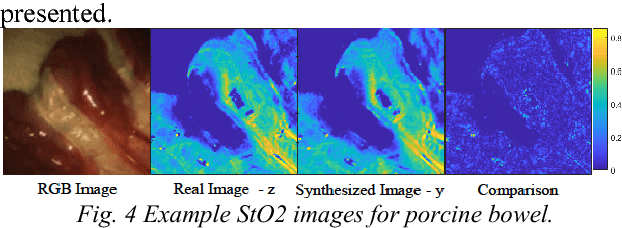
Abstract:Intra-operative measurement of tissue oxygen saturation (StO2) has been widely explored by pulse oximetry or hyperspectral imaging (HSI) to assess the function and viability of tissue. In this paper we propose a pixel- level image-to-image translation approach based on conditional Generative Adversarial Networks (cGAN) to estimate tissue oxygen saturation (StO2) directly from RGB images. The real-time performance and non-reliance on additional hardware, enable a seamless integration of the proposed method into surgical and diagnostic workflows with standard endoscope systems. For validation, RGB images and StO2 ground truth were simulated and estimated from HSI images collected by a liquid crystal tuneable filter (LCTF) endoscope for three tissue types (porcine bowel, lamb uterus and rabbit uterus). The result show that the proposed method can achieve visually identical images with comparable accuracy.
Augmented Reality needle ablation guidance tool for Irreversible Electroporation in the pancreas
Feb 09, 2018Abstract:Irreversible electroporation (IRE) is a soft tissue ablation technique suitable for treatment of inoperable tumours in the pancreas. The process involves applying a high voltage electric field to the tissue containing the mass using needle electrodes, leaving cancerous cells irreversibly damaged and vulnerable to apoptosis. Efficacy of the treatment depends heavily on the accuracy of needle placement and requires a high degree of skill from the operator. In this paper, we describe an Augmented Reality (AR) system designed to overcome the challenges associated with planning and guiding the needle insertion process. Our solution, based on the HoloLens (Microsoft, USA) platform, tracks the position of the headset, needle electrodes and ultrasound (US) probe in space. The proof of concept implementation of the system uses this tracking data to render real-time holographic guides on the HoloLens, giving the user insight into the current progress of needle insertion and an indication of the target needle trajectory. The operator's field of view is augmented using visual guides and real-time US feed rendered on a holographic plane, eliminating the need to consult external monitors. Based on these early prototypes, we are aiming to develop a system that will lower the skill level required for IRE while increasing overall accuracy of needle insertion and, hence, the likelihood of successful treatment.
Inference of Haemoglobin Concentration From Stereo RGB
Jun 22, 2017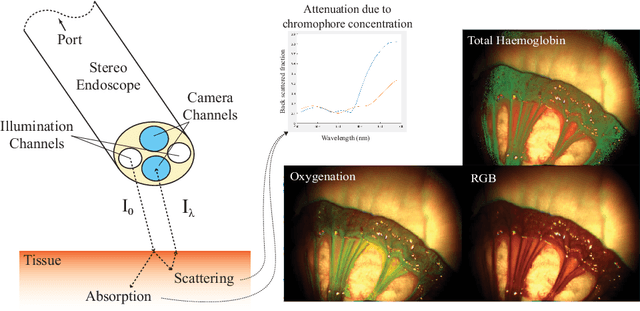

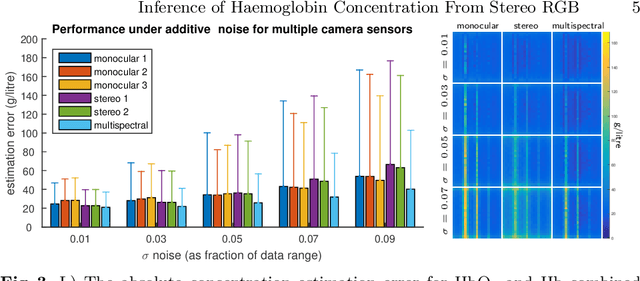

Abstract:Multispectral imaging (MSI) can provide information about tissue oxygenation, perfusion and potentially function during surgery. In this paper we present a novel, near real-time technique for intrinsic measurements of total haemoglobin (THb) and blood oxygenation (SO2) in tissue using only RGB images from a stereo laparoscope. The high degree of spectral overlap between channels makes inference of haemoglobin concentration challenging, non-linear and under constrained. We decompose the problem into two constrained linear sub-problems and show that with Tikhonov regularisation the estimation significantly improves, giving robust estimation of the Thb. We demonstrate by using the co-registered stereo image data from two cameras it is possible to get robust SO2 estimation as well. Our method is closed from, providing computational efficiency even with multiple cameras. The method we present requires only spectral response calibration of each camera, without modification of existing laparoscopic imaging hardware. We validate our technique on synthetic data from Monte Carlo simulation % of light transport through soft tissue containing submerged blood vessels and further, in vivo, on a multispectral porcine data set.
Endoscopic Depth Measurement and Super-Spectral-Resolution Imaging
Jun 21, 2017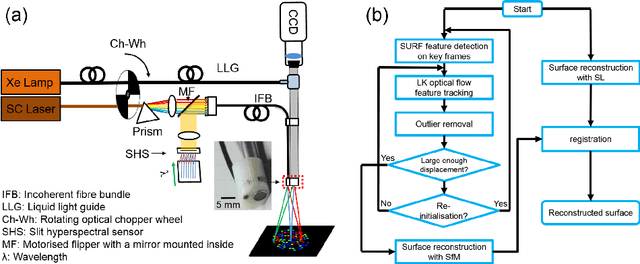

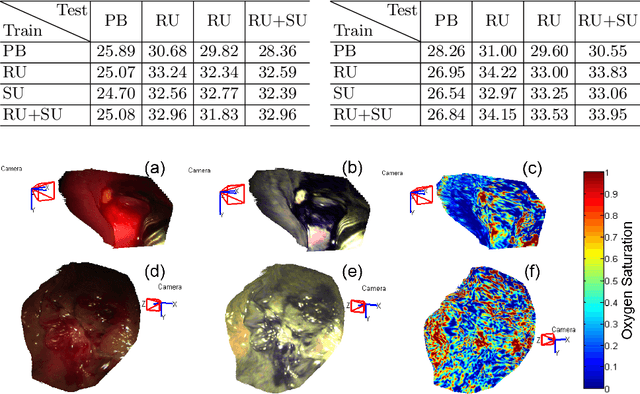
Abstract:Intra-operative measurements of tissue shape and multi/ hyperspectral information have the potential to provide surgical guidance and decision making support. We report an optical probe based system to combine sparse hyperspectral measurements and spectrally-encoded structured lighting (SL) for surface measurements. The system provides informative signals for navigation with a surgical interface. By rapidly switching between SL and white light (WL) modes, SL information is combined with structure-from-motion (SfM) from white light images, based on SURF feature detection and Lucas-Kanade (LK) optical flow to provide quasi-dense surface shape reconstruction with known scale in real-time. Furthermore, "super-spectral-resolution" was realized, whereby the RGB images and sparse hyperspectral data were integrated to recover dense pixel-level hyperspectral stacks, by using convolutional neural networks to upscale the wavelength dimension. Validation and demonstration of this system is reported on ex vivo/in vivo animal/ human experiments.
Recovering Dense Tissue Multispectral Signal from in vivo RGB Images
Jun 20, 2017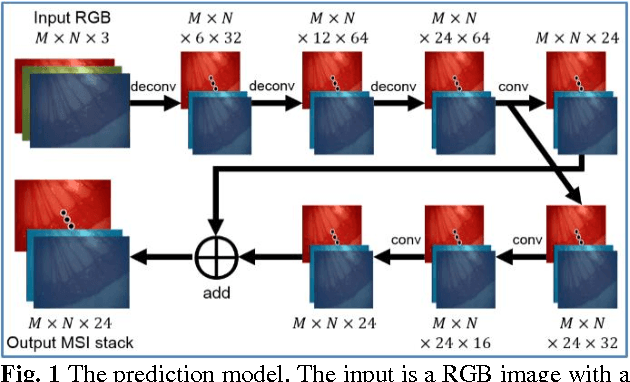
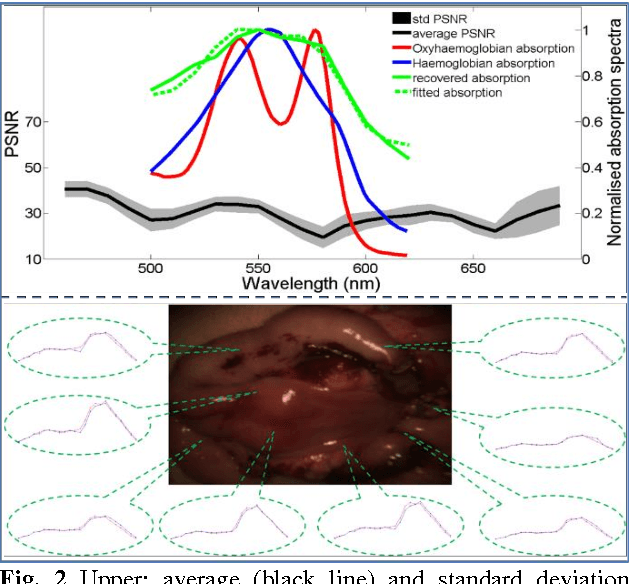
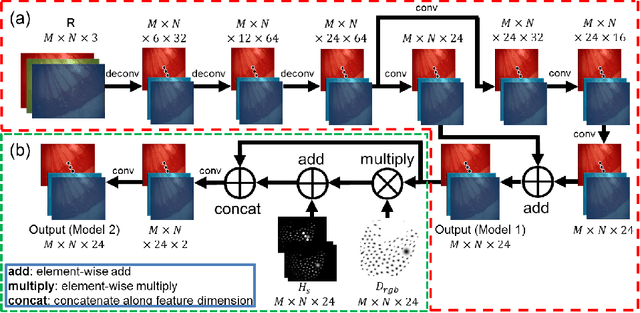
Abstract:Hyperspectral/multispectral imaging (HSI/MSI) contains rich information clinical applications, such as 1) narrow band imaging for vascular visualisation; 2) oxygen saturation for intraoperative perfusion monitoring and clinical decision making [1]; 3) tissue classification and identification of pathology [2]. The current systems which provide pixel-level HSI/MSI signal can be generally divided into two types: spatial scanning and spectral scanning. However, the trade-off between spatial/spectral resolution, the acquisition time, and the hardware complexity hampers implementation in real-world applications, especially intra-operatively. Acquiring high resolution images in real-time is important for HSI/MSI in intra-operative imaging, to alleviate the side effect caused by breathing, heartbeat, and other sources of motion. Therefore, we developed an algorithm to recover a pixel-level MSI stack using only the captured snapshot RGB images from a normal camera. We refer to this technique as "super-spectral-resolution". The proposed method enables recovery of pixel-level-dense MSI signals with 24 spectral bands at ~11 frames per second (FPS) on a GPU. Multispectral data captured from porcine bowel and sheep/rabbit uteri in vivo has been used for training, and the algorithm has been validated using unseen in vivo animal experiments.
Probe-based Rapid Hybrid Hyperspectral and Tissue Surface Imaging Aided by Fully Convolutional Networks
Jun 15, 2016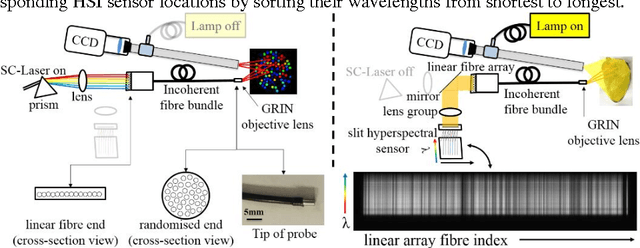
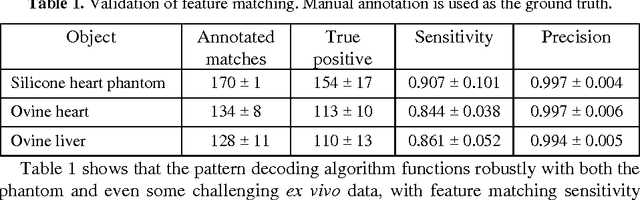
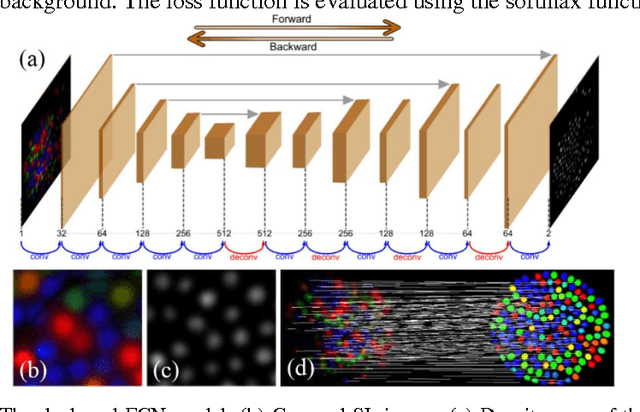
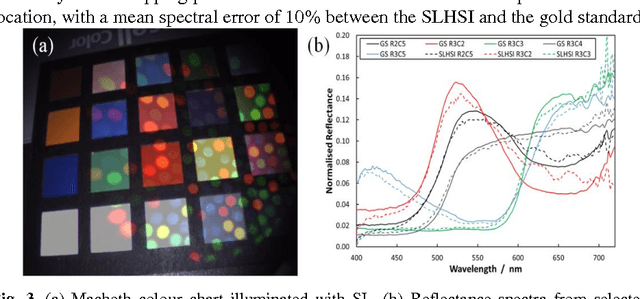
Abstract:Tissue surface shape and reflectance spectra provide rich intra-operative information useful in surgical guidance. We propose a hybrid system which displays an endoscopic image with a fast joint inspection of tissue surface shape using structured light (SL) and hyperspectral imaging (HSI). For SL a miniature fibre probe is used to project a coloured spot pattern onto the tissue surface. In HSI mode standard endoscopic illumination is used, with the fibre probe collecting reflected light and encoding the spatial information into a linear format that can be imaged onto the slit of a spectrograph. Correspondence between the arrangement of fibres at the distal and proximal ends of the bundle was found using spectral encoding. Then during pattern decoding, a fully convolutional network (FCN) was used for spot detection, followed by a matching propagation algorithm for spot identification. This method enabled fast reconstruction (12 frames per second) using a GPU. The hyperspectral image was combined with the white light image and the reconstructed surface, showing the spectral information of different areas. Validation of this system using phantom and ex vivo experiments has been demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge