Hui Cui
Iterative pseudo-labeling based adaptive copy-paste supervision for semi-supervised tumor segmentation
Aug 06, 2025Abstract:Semi-supervised learning (SSL) has attracted considerable attention in medical image processing. The latest SSL methods use a combination of consistency regularization and pseudo-labeling to achieve remarkable success. However, most existing SSL studies focus on segmenting large organs, neglecting the challenging scenarios where there are numerous tumors or tumors of small volume. Furthermore, the extensive capabilities of data augmentation strategies, particularly in the context of both labeled and unlabeled data, have yet to be thoroughly investigated. To tackle these challenges, we introduce a straightforward yet effective approach, termed iterative pseudo-labeling based adaptive copy-paste supervision (IPA-CP), for tumor segmentation in CT scans. IPA-CP incorporates a two-way uncertainty based adaptive augmentation mechanism, aiming to inject tumor uncertainties present in the mean teacher architecture into adaptive augmentation. Additionally, IPA-CP employs an iterative pseudo-label transition strategy to generate more robust and informative pseudo labels for the unlabeled samples. Extensive experiments on both in-house and public datasets show that our framework outperforms state-of-the-art SSL methods in medical image segmentation. Ablation study results demonstrate the effectiveness of our technical contributions.
TSEML: A task-specific embedding-based method for few-shot classification of cancer molecular subtypes
Dec 17, 2024Abstract:Molecular subtyping of cancer is recognized as a critical and challenging upstream task for personalized therapy. Existing deep learning methods have achieved significant performance in this domain when abundant data samples are available. However, the acquisition of densely labeled samples for cancer molecular subtypes remains a significant challenge for conventional data-intensive deep learning approaches. In this work, we focus on the few-shot molecular subtype prediction problem in heterogeneous and small cancer datasets, aiming to enhance precise diagnosis and personalized treatment. We first construct a new few-shot dataset for cancer molecular subtype classification and auxiliary cancer classification, named TCGA Few-Shot, from existing publicly available datasets. To effectively leverage the relevant knowledge from both tasks, we introduce a task-specific embedding-based meta-learning framework (TSEML). TSEML leverages the synergistic strengths of a model-agnostic meta-learning (MAML) approach and a prototypical network (ProtoNet) to capture diverse and fine-grained features. Comparative experiments conducted on the TCGA Few-Shot dataset demonstrate that our TSEML framework achieves superior performance in addressing the problem of few-shot molecular subtype classification.
Location embedding based pairwise distance learning for fine-grained diagnosis of urinary stones
Jun 29, 2024Abstract:The precise diagnosis of urinary stones is crucial for devising effective treatment strategies. The diagnostic process, however, is often complicated by the low contrast between stones and surrounding tissues, as well as the variability in stone locations across different patients. To address this issue, we propose a novel location embedding based pairwise distance learning network (LEPD-Net) that leverages low-dose abdominal X-ray imaging combined with location information for the fine-grained diagnosis of urinary stones. LEPD-Net enhances the representation of stone-related features through context-aware region enhancement, incorporates critical location knowledge via stone location embedding, and achieves recognition of fine-grained objects with our innovative fine-grained pairwise distance learning. Additionally, we have established an in-house dataset on urinary tract stones to demonstrate the effectiveness of our proposed approach. Comprehensive experiments conducted on this dataset reveal that our framework significantly surpasses existing state-of-the-art methods.
Inter- and intra-uncertainty based feature aggregation model for semi-supervised histopathology image segmentation
Mar 19, 2024Abstract:Acquiring pixel-level annotations is often limited in applications such as histology studies that require domain expertise. Various semi-supervised learning approaches have been developed to work with limited ground truth annotations, such as the popular teacher-student models. However, hierarchical prediction uncertainty within the student model (intra-uncertainty) and image prediction uncertainty (inter-uncertainty) have not been fully utilized by existing methods. To address these issues, we first propose a novel inter- and intra-uncertainty regularization method to measure and constrain both inter- and intra-inconsistencies in the teacher-student architecture. We also propose a new two-stage network with pseudo-mask guided feature aggregation (PG-FANet) as the segmentation model. The two-stage structure complements with the uncertainty regularization strategy to avoid introducing extra modules in solving uncertainties and the aggregation mechanisms enable multi-scale and multi-stage feature integration. Comprehensive experimental results over the MoNuSeg and CRAG datasets show that our PG-FANet outperforms other state-of-the-art methods and our semi-supervised learning framework yields competitive performance with a limited amount of labeled data.
MGCT: Mutual-Guided Cross-Modality Transformer for Survival Outcome Prediction using Integrative Histopathology-Genomic Features
Nov 20, 2023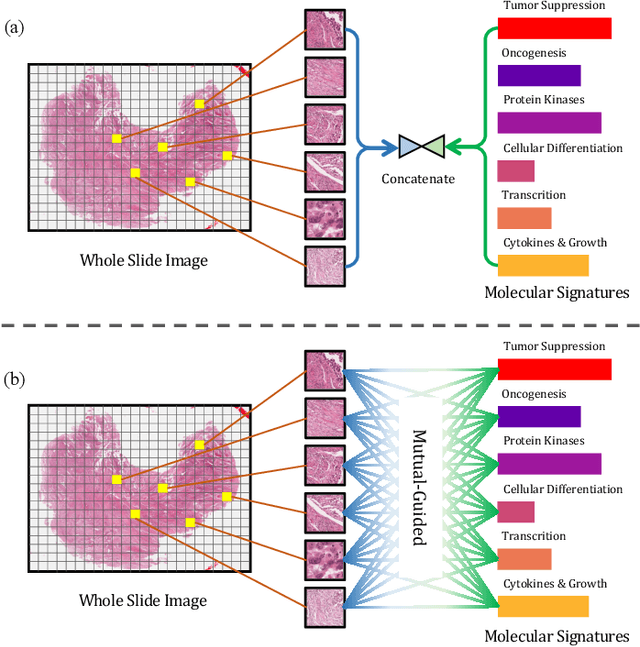
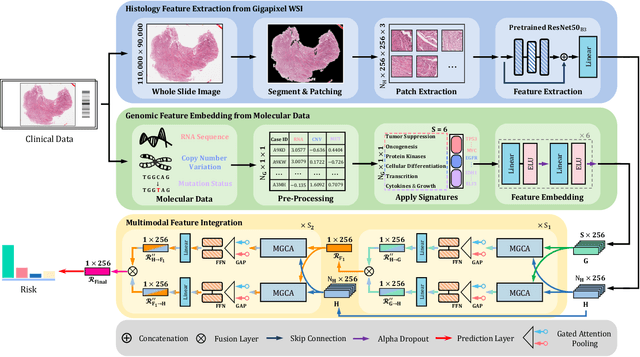
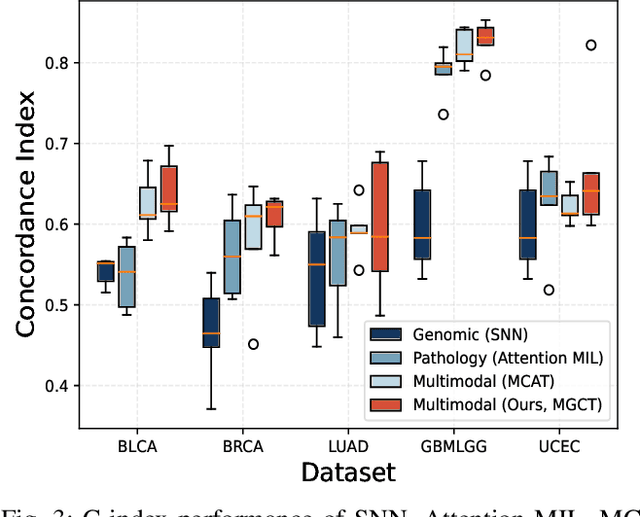
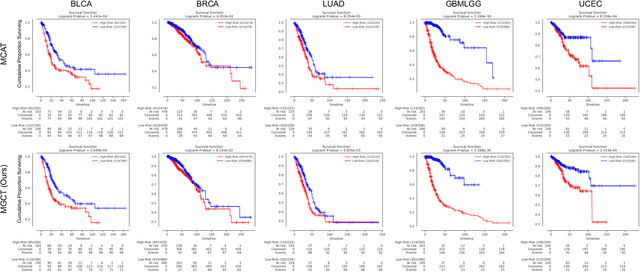
Abstract:The rapidly emerging field of deep learning-based computational pathology has shown promising results in utilizing whole slide images (WSIs) to objectively prognosticate cancer patients. However, most prognostic methods are currently limited to either histopathology or genomics alone, which inevitably reduces their potential to accurately predict patient prognosis. Whereas integrating WSIs and genomic features presents three main challenges: (1) the enormous heterogeneity of gigapixel WSIs which can reach sizes as large as 150,000x150,000 pixels; (2) the absence of a spatially corresponding relationship between histopathology images and genomic molecular data; and (3) the existing early, late, and intermediate multimodal feature fusion strategies struggle to capture the explicit interactions between WSIs and genomics. To ameliorate these issues, we propose the Mutual-Guided Cross-Modality Transformer (MGCT), a weakly-supervised, attention-based multimodal learning framework that can combine histology features and genomic features to model the genotype-phenotype interactions within the tumor microenvironment. To validate the effectiveness of MGCT, we conduct experiments using nearly 3,600 gigapixel WSIs across five different cancer types sourced from The Cancer Genome Atlas (TCGA). Extensive experimental results consistently emphasize that MGCT outperforms the state-of-the-art (SOTA) methods.
Free-form tumor synthesis in computed tomography images via richer generative adversarial network
Apr 20, 2021
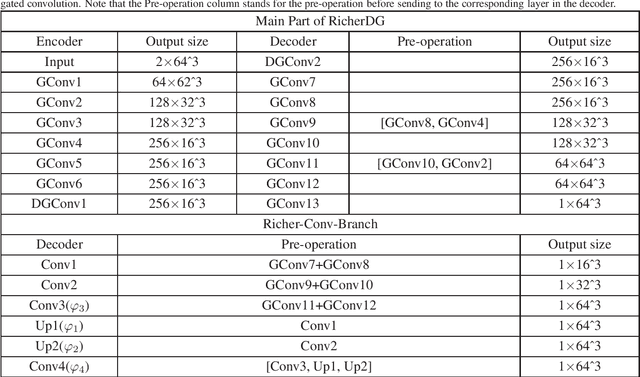

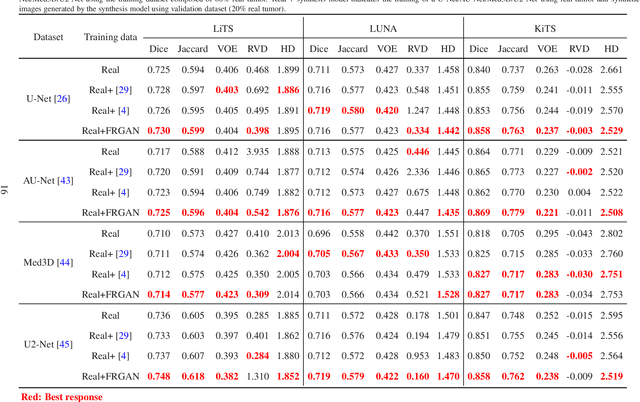
Abstract:The insufficiency of annotated medical imaging scans for cancer makes it challenging to train and validate data-hungry deep learning models in precision oncology. We propose a new richer generative adversarial network for free-form 3D tumor/lesion synthesis in computed tomography (CT) images. The network is composed of a new richer convolutional feature enhanced dilated-gated generator (RicherDG) and a hybrid loss function. The RicherDG has dilated-gated convolution layers to enable tumor-painting and to enlarge perceptive fields; and it has a novel richer convolutional feature association branch to recover multi-scale convolutional features especially from uncertain boundaries between tumor and surrounding healthy tissues. The hybrid loss function, which consists of a diverse range of losses, is designed to aggregate complementary information to improve optimization. We perform a comprehensive evaluation of the synthesis results on a wide range of public CT image datasets covering the liver, kidney tumors, and lung nodules. The qualitative and quantitative evaluations and ablation study demonstrated improved synthesizing results over advanced tumor synthesis methods.
Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images
Apr 20, 2021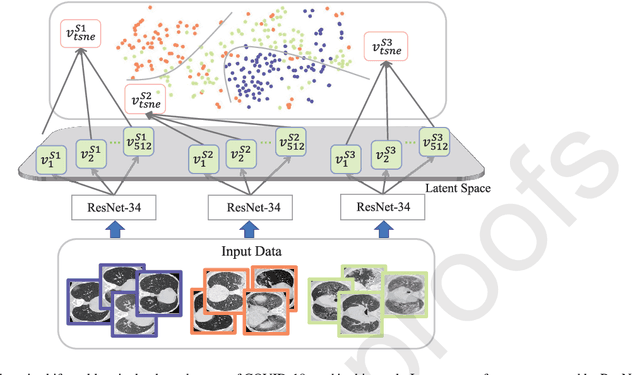
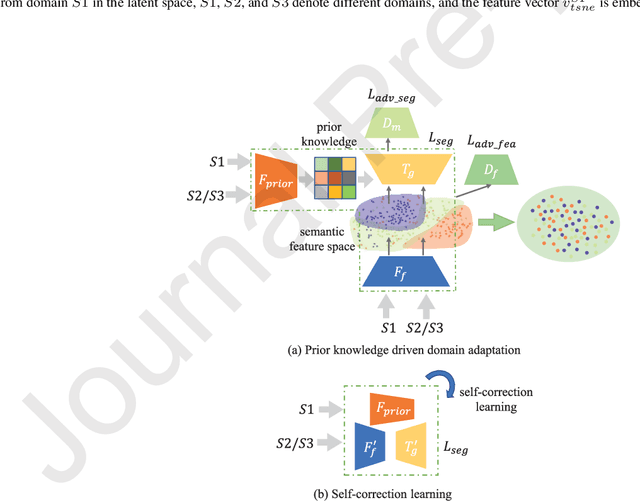
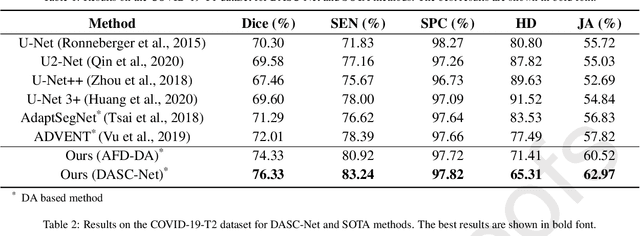
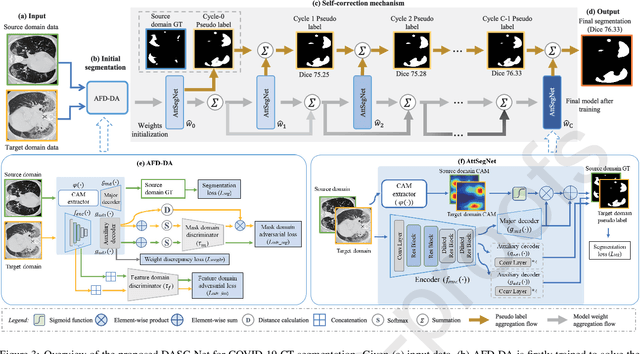
Abstract:The capability of generalization to unseen domains is crucial for deep learning models when considering real-world scenarios. However, current available medical image datasets, such as those for COVID-19 CT images, have large variations of infections and domain shift problems. To address this issue, we propose a prior knowledge driven domain adaptation and a dual-domain enhanced self-correction learning scheme. Based on the novel learning schemes, a domain adaptation based self-correction model (DASC-Net) is proposed for COVID-19 infection segmentation on CT images. DASC-Net consists of a novel attention and feature domain enhanced domain adaptation model (AFD-DA) to solve the domain shifts and a self-correction learning process to refine segmentation results. The innovations in AFD-DA include an image-level activation feature extractor with attention to lung abnormalities and a multi-level discrimination module for hierarchical feature domain alignment. The proposed self-correction learning process adaptively aggregates the learned model and corresponding pseudo labels for the propagation of aligned source and target domain information to alleviate the overfitting to noises caused by pseudo labels. Extensive experiments over three publicly available COVID-19 CT datasets demonstrate that DASC-Net consistently outperforms state-of-the-art segmentation, domain shift, and coronavirus infection segmentation methods. Ablation analysis further shows the effectiveness of the major components in our model. The DASC-Net enriches the theory of domain adaptation and self-correction learning in medical imaging and can be generalized to multi-site COVID-19 infection segmentation on CT images for clinical deployment.
SigNet: An Advanced Deep Learning Framework for Radio Signal Classification
Oct 28, 2020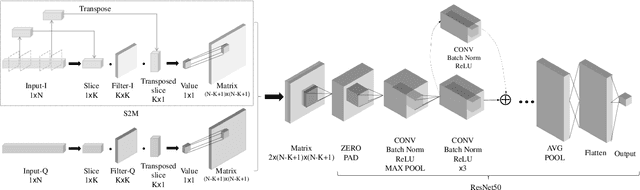
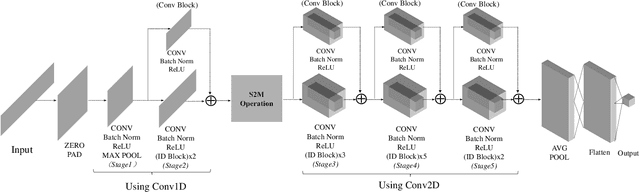
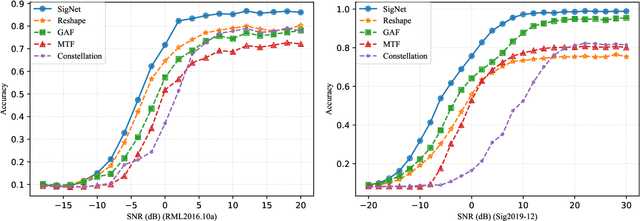
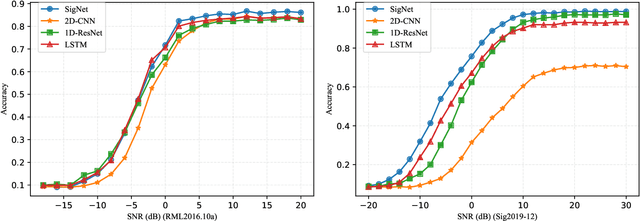
Abstract:Deep learning methods achieve great success in many areas due to their powerful feature extraction capabilities and end-to-end training mechanism, and recently they are also introduced for radio signal modulation classification. In this paper, we propose a novel deep learning framework called SigNet, where a signal-to-matrix (S2M) operator is adopted to convert the original signal into a square matrix first and is co-trained with a follow-up CNN architecture for classification. This model is further accelerated by integrating 1D convolution operators, leading to the upgraded model SigNet2.0. The experiments on two signal datasets show that both SigNet and SigNet2.0 outperform a number of well-known baselines, achieving the state-of-the-art performance. Notably, they obtain significantly higher accuracy than 1D-ResNet and 2D-CNN (at most increasing 70.5\%), while much faster than LSTM (at most saving 88.0\% training time). More interestingly, our proposed models behave extremely well in few-shot learning when a small training data set is provided. They can achieve a relatively high accuracy even when 1\% training data are kept, while other baseline models may lose their effectiveness much more quickly as the datasets get smaller. Such result suggests that SigNet/SigNet2.0 could be extremely useful in the situations where labeled signal data are difficult to obtain.
Dual-level Semantic Transfer Deep Hashing for Efficient Social Image Retrieval
Jun 10, 2020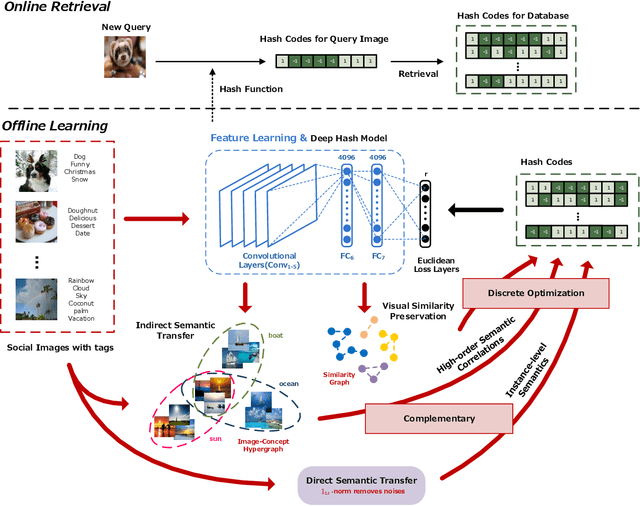
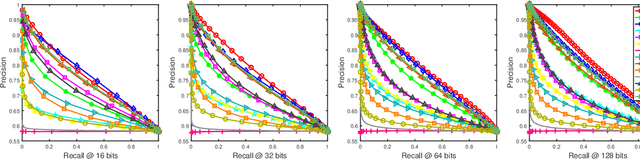
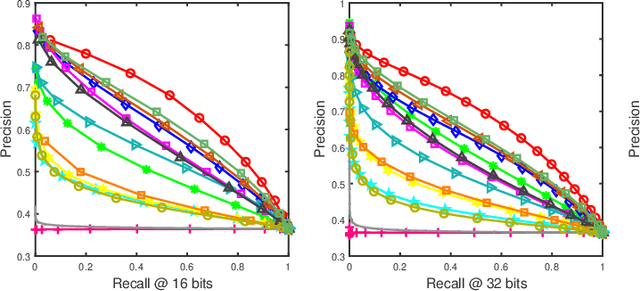
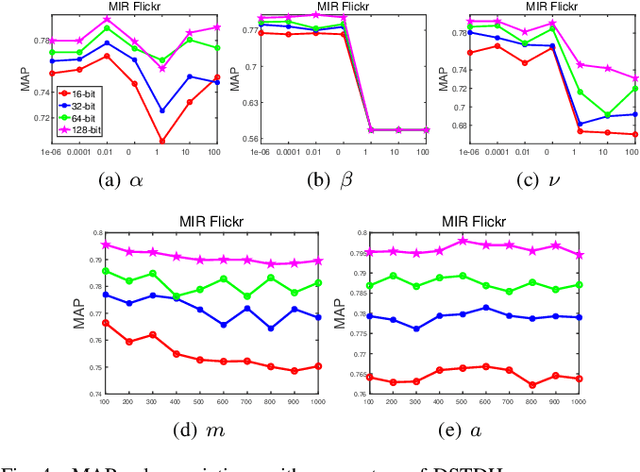
Abstract:Social network stores and disseminates a tremendous amount of user shared images. Deep hashing is an efficient indexing technique to support large-scale social image retrieval, due to its deep representation capability, fast retrieval speed and low storage cost. Particularly, unsupervised deep hashing has well scalability as it does not require any manually labelled data for training. However, owing to the lacking of label guidance, existing methods suffer from severe semantic shortage when optimizing a large amount of deep neural network parameters. Differently, in this paper, we propose a Dual-level Semantic Transfer Deep Hashing (DSTDH) method to alleviate this problem with a unified deep hash learning framework. Our model targets at learning the semantically enhanced deep hash codes by specially exploiting the user-generated tags associated with the social images. Specifically, we design a complementary dual-level semantic transfer mechanism to efficiently discover the potential semantics of tags and seamlessly transfer them into binary hash codes. On the one hand, instance-level semantics are directly preserved into hash codes from the associated tags with adverse noise removing. Besides, an image-concept hypergraph is constructed for indirectly transferring the latent high-order semantic correlations of images and tags into hash codes. Moreover, the hash codes are obtained simultaneously with the deep representation learning by the discrete hash optimization strategy. Extensive experiments on two public social image retrieval datasets validate the superior performance of our method compared with state-of-the-art hashing methods. The source codes of our method can be obtained at https://github.com/research2020-1/DSTDH
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge