Qiangguo Jin
ERSR: An Ellipse-constrained pseudo-label refinement and symmetric regularization framework for semi-supervised fetal head segmentation in ultrasound images
Aug 27, 2025
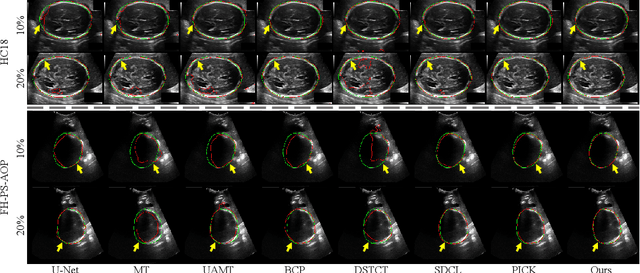

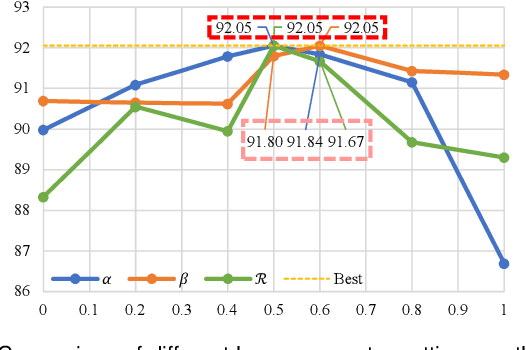
Abstract:Automated segmentation of the fetal head in ultrasound images is critical for prenatal monitoring. However, achieving robust segmentation remains challenging due to the poor quality of ultrasound images and the lack of annotated data. Semi-supervised methods alleviate the lack of annotated data but struggle with the unique characteristics of fetal head ultrasound images, making it challenging to generate reliable pseudo-labels and enforce effective consistency regularization constraints. To address this issue, we propose a novel semi-supervised framework, ERSR, for fetal head ultrasound segmentation. Our framework consists of the dual-scoring adaptive filtering strategy, the ellipse-constrained pseudo-label refinement, and the symmetry-based multiple consistency regularization. The dual-scoring adaptive filtering strategy uses boundary consistency and contour regularity criteria to evaluate and filter teacher outputs. The ellipse-constrained pseudo-label refinement refines these filtered outputs by fitting least-squares ellipses, which strengthens pixels near the center of the fitted ellipse and suppresses noise simultaneously. The symmetry-based multiple consistency regularization enforces multi-level consistency across perturbed images, symmetric regions, and between original predictions and pseudo-labels, enabling the model to capture robust and stable shape representations. Our method achieves state-of-the-art performance on two benchmarks. On the HC18 dataset, it reaches Dice scores of 92.05% and 95.36% with 10% and 20% labeled data, respectively. On the PSFH dataset, the scores are 91.68% and 93.70% under the same settings.
Iterative pseudo-labeling based adaptive copy-paste supervision for semi-supervised tumor segmentation
Aug 06, 2025Abstract:Semi-supervised learning (SSL) has attracted considerable attention in medical image processing. The latest SSL methods use a combination of consistency regularization and pseudo-labeling to achieve remarkable success. However, most existing SSL studies focus on segmenting large organs, neglecting the challenging scenarios where there are numerous tumors or tumors of small volume. Furthermore, the extensive capabilities of data augmentation strategies, particularly in the context of both labeled and unlabeled data, have yet to be thoroughly investigated. To tackle these challenges, we introduce a straightforward yet effective approach, termed iterative pseudo-labeling based adaptive copy-paste supervision (IPA-CP), for tumor segmentation in CT scans. IPA-CP incorporates a two-way uncertainty based adaptive augmentation mechanism, aiming to inject tumor uncertainties present in the mean teacher architecture into adaptive augmentation. Additionally, IPA-CP employs an iterative pseudo-label transition strategy to generate more robust and informative pseudo labels for the unlabeled samples. Extensive experiments on both in-house and public datasets show that our framework outperforms state-of-the-art SSL methods in medical image segmentation. Ablation study results demonstrate the effectiveness of our technical contributions.
TSEML: A task-specific embedding-based method for few-shot classification of cancer molecular subtypes
Dec 17, 2024Abstract:Molecular subtyping of cancer is recognized as a critical and challenging upstream task for personalized therapy. Existing deep learning methods have achieved significant performance in this domain when abundant data samples are available. However, the acquisition of densely labeled samples for cancer molecular subtypes remains a significant challenge for conventional data-intensive deep learning approaches. In this work, we focus on the few-shot molecular subtype prediction problem in heterogeneous and small cancer datasets, aiming to enhance precise diagnosis and personalized treatment. We first construct a new few-shot dataset for cancer molecular subtype classification and auxiliary cancer classification, named TCGA Few-Shot, from existing publicly available datasets. To effectively leverage the relevant knowledge from both tasks, we introduce a task-specific embedding-based meta-learning framework (TSEML). TSEML leverages the synergistic strengths of a model-agnostic meta-learning (MAML) approach and a prototypical network (ProtoNet) to capture diverse and fine-grained features. Comparative experiments conducted on the TCGA Few-Shot dataset demonstrate that our TSEML framework achieves superior performance in addressing the problem of few-shot molecular subtype classification.
Location embedding based pairwise distance learning for fine-grained diagnosis of urinary stones
Jun 29, 2024Abstract:The precise diagnosis of urinary stones is crucial for devising effective treatment strategies. The diagnostic process, however, is often complicated by the low contrast between stones and surrounding tissues, as well as the variability in stone locations across different patients. To address this issue, we propose a novel location embedding based pairwise distance learning network (LEPD-Net) that leverages low-dose abdominal X-ray imaging combined with location information for the fine-grained diagnosis of urinary stones. LEPD-Net enhances the representation of stone-related features through context-aware region enhancement, incorporates critical location knowledge via stone location embedding, and achieves recognition of fine-grained objects with our innovative fine-grained pairwise distance learning. Additionally, we have established an in-house dataset on urinary tract stones to demonstrate the effectiveness of our proposed approach. Comprehensive experiments conducted on this dataset reveal that our framework significantly surpasses existing state-of-the-art methods.
Inter- and intra-uncertainty based feature aggregation model for semi-supervised histopathology image segmentation
Mar 19, 2024Abstract:Acquiring pixel-level annotations is often limited in applications such as histology studies that require domain expertise. Various semi-supervised learning approaches have been developed to work with limited ground truth annotations, such as the popular teacher-student models. However, hierarchical prediction uncertainty within the student model (intra-uncertainty) and image prediction uncertainty (inter-uncertainty) have not been fully utilized by existing methods. To address these issues, we first propose a novel inter- and intra-uncertainty regularization method to measure and constrain both inter- and intra-inconsistencies in the teacher-student architecture. We also propose a new two-stage network with pseudo-mask guided feature aggregation (PG-FANet) as the segmentation model. The two-stage structure complements with the uncertainty regularization strategy to avoid introducing extra modules in solving uncertainties and the aggregation mechanisms enable multi-scale and multi-stage feature integration. Comprehensive experimental results over the MoNuSeg and CRAG datasets show that our PG-FANet outperforms other state-of-the-art methods and our semi-supervised learning framework yields competitive performance with a limited amount of labeled data.
Prototype as Query for Few Shot Semantic Segmentation
Nov 27, 2022



Abstract:Few-shot Semantic Segmentation (FSS) was proposed to segment unseen classes in a query image, referring to only a few annotated examples named support images. One of the characteristics of FSS is spatial inconsistency between query and support targets, e.g., texture or appearance. This greatly challenges the generalization ability of methods for FSS, which requires to effectively exploit the dependency of the query image and the support examples. Most existing methods abstracted support features into prototype vectors and implemented the interaction with query features using cosine similarity or feature concatenation. However, this simple interaction may not capture spatial details in query features. To alleviate this limitation, a few methods utilized all pixel-wise support information via computing the pixel-wise correlations between paired query and support features implemented with the attention mechanism of Transformer. These approaches suffer from heavy computation on the dot-product attention between all pixels of support and query features. In this paper, we propose a simple yet effective framework built upon Transformer termed as ProtoFormer to fully capture spatial details in query features. It views the abstracted prototype of the target class in support features as Query and the query features as Key and Value embeddings, which are input to the Transformer decoder. In this way, the spatial details can be better captured and the semantic features of target class in the query image can be focused. The output of the Transformer-based module can be viewed as semantic-aware dynamic kernels to filter out the segmentation mask from the enriched query features. Extensive experiments on PASCAL-$5^{i}$ and COCO-$20^{i}$ show that our ProtoFormer significantly advances the state-of-the-art methods.
Free-form tumor synthesis in computed tomography images via richer generative adversarial network
Apr 20, 2021
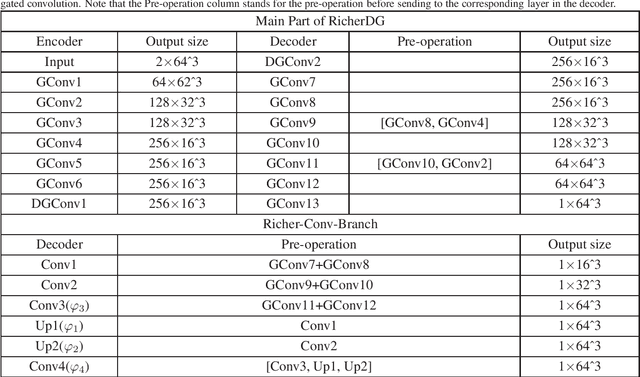

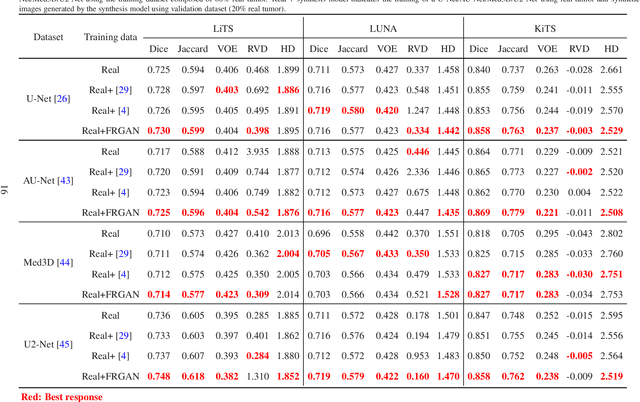
Abstract:The insufficiency of annotated medical imaging scans for cancer makes it challenging to train and validate data-hungry deep learning models in precision oncology. We propose a new richer generative adversarial network for free-form 3D tumor/lesion synthesis in computed tomography (CT) images. The network is composed of a new richer convolutional feature enhanced dilated-gated generator (RicherDG) and a hybrid loss function. The RicherDG has dilated-gated convolution layers to enable tumor-painting and to enlarge perceptive fields; and it has a novel richer convolutional feature association branch to recover multi-scale convolutional features especially from uncertain boundaries between tumor and surrounding healthy tissues. The hybrid loss function, which consists of a diverse range of losses, is designed to aggregate complementary information to improve optimization. We perform a comprehensive evaluation of the synthesis results on a wide range of public CT image datasets covering the liver, kidney tumors, and lung nodules. The qualitative and quantitative evaluations and ablation study demonstrated improved synthesizing results over advanced tumor synthesis methods.
Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images
Apr 20, 2021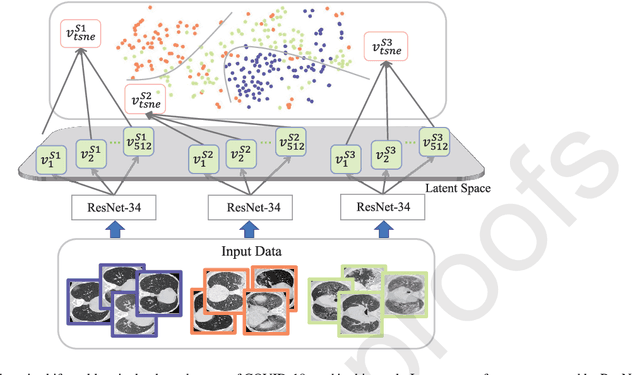
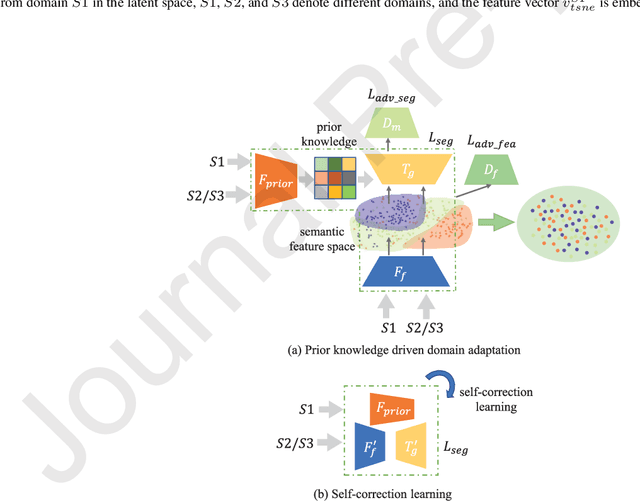
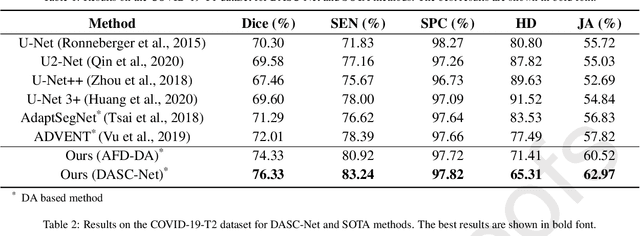
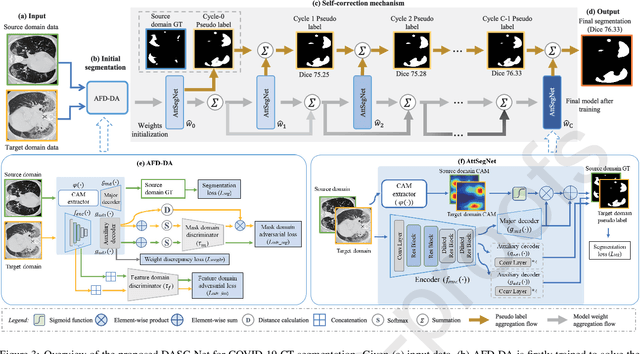
Abstract:The capability of generalization to unseen domains is crucial for deep learning models when considering real-world scenarios. However, current available medical image datasets, such as those for COVID-19 CT images, have large variations of infections and domain shift problems. To address this issue, we propose a prior knowledge driven domain adaptation and a dual-domain enhanced self-correction learning scheme. Based on the novel learning schemes, a domain adaptation based self-correction model (DASC-Net) is proposed for COVID-19 infection segmentation on CT images. DASC-Net consists of a novel attention and feature domain enhanced domain adaptation model (AFD-DA) to solve the domain shifts and a self-correction learning process to refine segmentation results. The innovations in AFD-DA include an image-level activation feature extractor with attention to lung abnormalities and a multi-level discrimination module for hierarchical feature domain alignment. The proposed self-correction learning process adaptively aggregates the learned model and corresponding pseudo labels for the propagation of aligned source and target domain information to alleviate the overfitting to noises caused by pseudo labels. Extensive experiments over three publicly available COVID-19 CT datasets demonstrate that DASC-Net consistently outperforms state-of-the-art segmentation, domain shift, and coronavirus infection segmentation methods. Ablation analysis further shows the effectiveness of the major components in our model. The DASC-Net enriches the theory of domain adaptation and self-correction learning in medical imaging and can be generalized to multi-site COVID-19 infection segmentation on CT images for clinical deployment.
RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans
Nov 04, 2018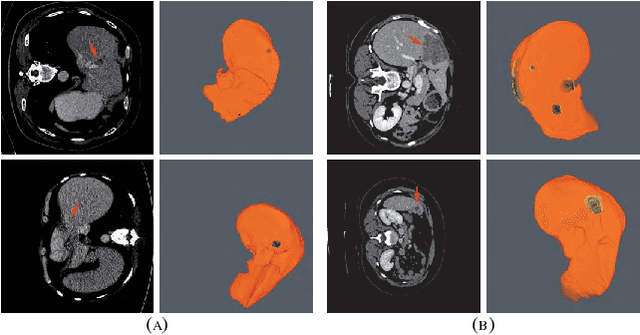
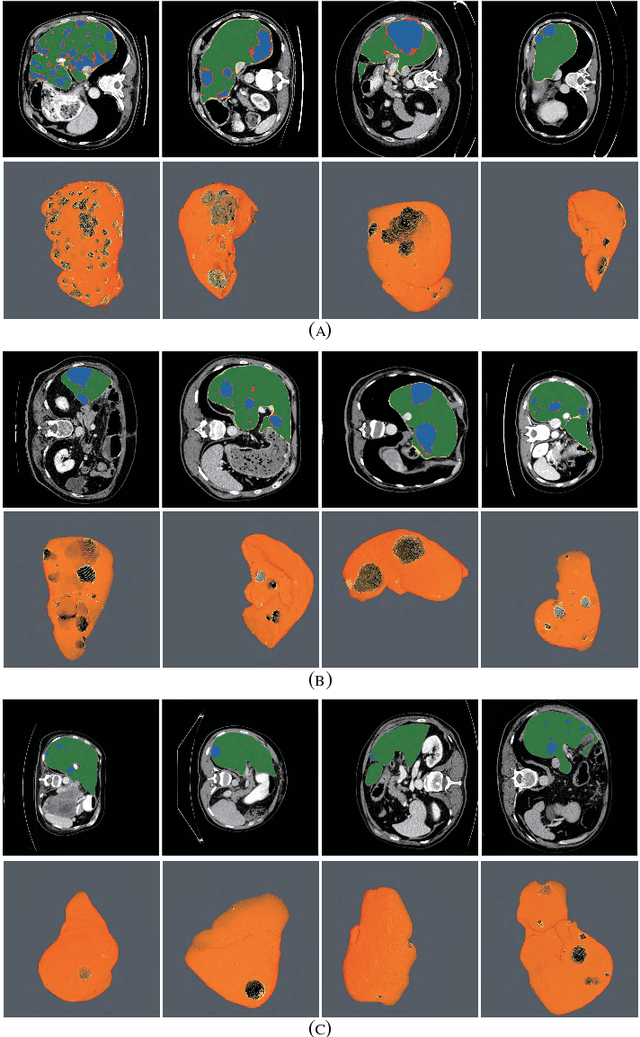
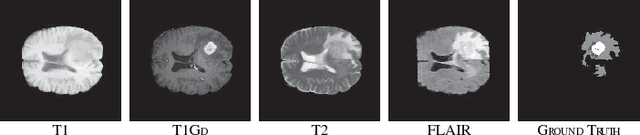

Abstract:Automatic extraction of liver and tumor from CT volumes is a challenging task due to their heterogeneous and diffusive shapes. Recently, 2D and 3D deep convolutional neural networks have become popular in medical image segmentation tasks because of the utilization of large labeled datasets to learn hierarchical features. However, 3D networks have some drawbacks due to their high cost on computational resources. In this paper, we propose a 3D hybrid residual attention-aware segmentation method, named RA-UNet, to precisely extract the liver volume of interests (VOI) and segment tumors from the liver VOI. The proposed network has a basic architecture as a 3D U-Net which extracts contextual information combining low-level feature maps with high-level ones. Attention modules are stacked so that the attention-aware features change adaptively as the network goes "very deep" and this is made possible by residual learning. This is the first work that an attention residual mechanism is used to process medical volumetric images. We evaluated our framework on the public MICCAI 2017 Liver Tumor Segmentation dataset and the 3DIRCADb dataset. The results show that our architecture outperforms other state-of-the-art methods. We also extend our RA-UNet to brain tumor segmentation on the BraTS2018 and BraTS2017 datasets, and the results indicate that RA-UNet achieves good performance on a brain tumor segmentation task as well.
DUNet: A deformable network for retinal vessel segmentation
Nov 03, 2018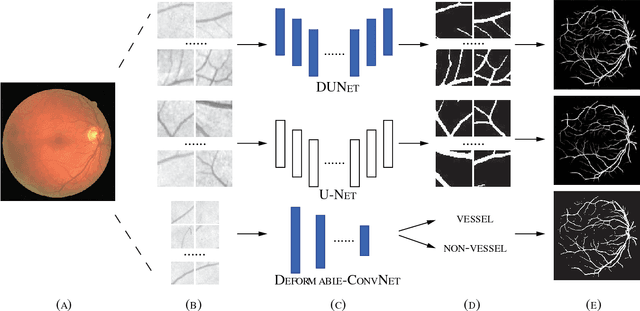
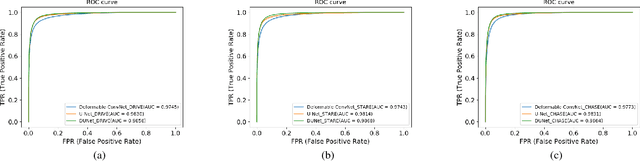
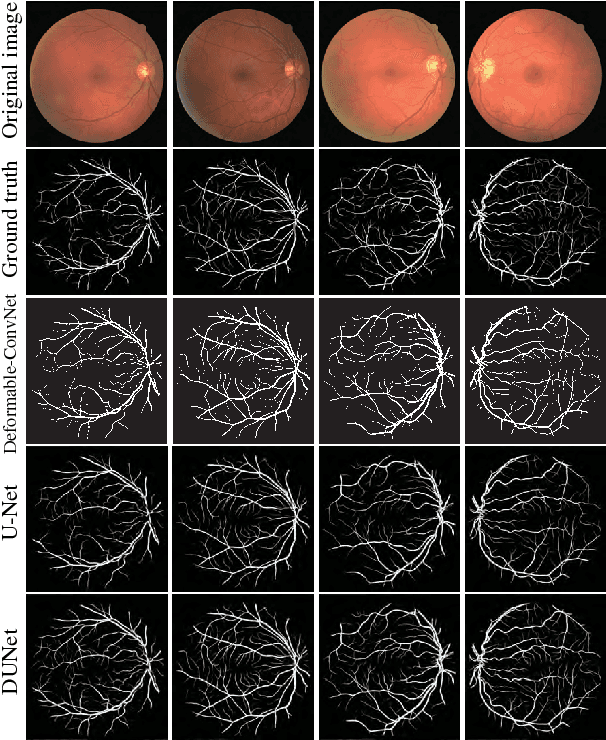
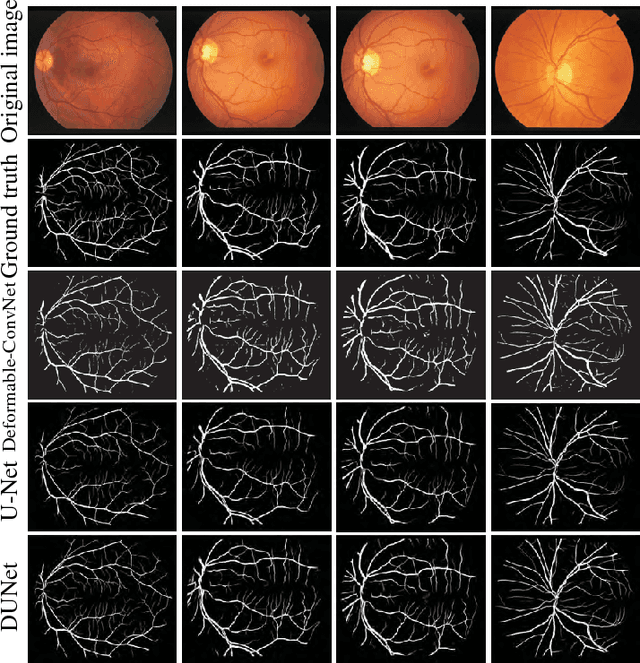
Abstract:Automatic segmentation of retinal vessels in fundus images plays an important role in the diagnosis of some diseases such as diabetes and hypertension. In this paper, we propose Deformable U-Net (DUNet), which exploits the retinal vessels' local features with a U-shape architecture, in an end to end manner for retinal vessel segmentation. Inspired by the recently introduced deformable convolutional networks, we integrate the deformable convolution into the proposed network. The DUNet, with upsampling operators to increase the output resolution, is designed to extract context information and enable precise localization by combining low-level feature maps with high-level ones. Furthermore, DUNet captures the retinal vessels at various shapes and scales by adaptively adjusting the receptive fields according to vessels' scales and shapes. Three public datasets DRIVE, STARE and CHASE_DB1 are used to train and test our model. Detailed comparisons between the proposed network and the deformable neural network, U-Net are provided in our study. Results show that more detailed vessels are extracted by DUNet and it exhibits state-of-the-art performance for retinal vessel segmentation with a global accuracy of 0.9697/0.9722/0.9724 and AUC of 0.9856/0.9868/0.9863 on DRIVE, STARE and CHASE_DB1 respectively. Moreover, to show the generalization ability of the DUNet, we used another two retinal vessel data sets, one is named WIDE and the other is a synthetic data set with diverse styles, named SYNTHE, to qualitatively and quantitatively analyzed and compared with other methods. Results indicates that DUNet outperforms other state-of-the-arts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge