Chunquan Li
Uncertainty-driven and Adversarial Calibration Learning for Epicardial Adipose Tissue Segmentation
Feb 23, 2024
Abstract:Epicardial adipose tissue (EAT) is a type of visceral fat that can secrete large amounts of adipokines to affect the myocardium and coronary arteries. EAT volume and density can be used as independent risk markers measurement of volume by noninvasive magnetic resonance images is the best method of assessing EAT. However, segmenting EAT is challenging due to the low contrast between EAT and pericardial effusion and the presence of motion artifacts. we propose a novel feature latent space multilevel supervision network (SPDNet) with uncertainty-driven and adversarial calibration learning to enhance segmentation for more accurate EAT volume estimation. The network first addresses the blurring of EAT edges due to the medical images in the open medical environments with low quality or out-of-distribution by modeling the uncertainty as a Gaussian distribution in the feature latent space, which using its Bayesian estimation as a regularization constraint to optimize SwinUNETR. Second, an adversarial training strategy is introduced to calibrate the segmentation feature map and consider the multi-scale feature differences between the uncertainty-guided predictive segmentation and the ground truth segmentation, synthesizing the multi-scale adversarial loss directly improves the ability to discriminate the similarity between organizations. Experiments on both the cardiac public MRI dataset (ACDC) and the real-world clinical cohort EAT dataset show that the proposed network outperforms mainstream models, validating that uncertainty-driven and adversarial calibration learning can be used to provide additional information for modeling multi-scale ambiguities.
MGCT: Mutual-Guided Cross-Modality Transformer for Survival Outcome Prediction using Integrative Histopathology-Genomic Features
Nov 20, 2023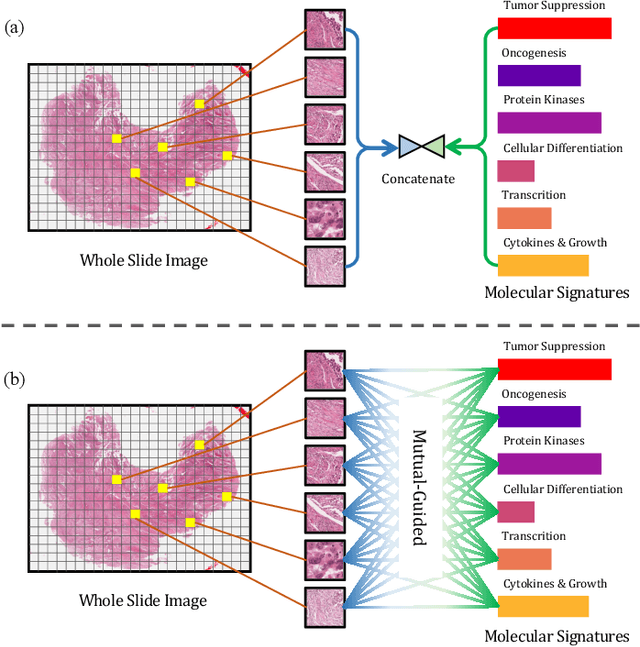
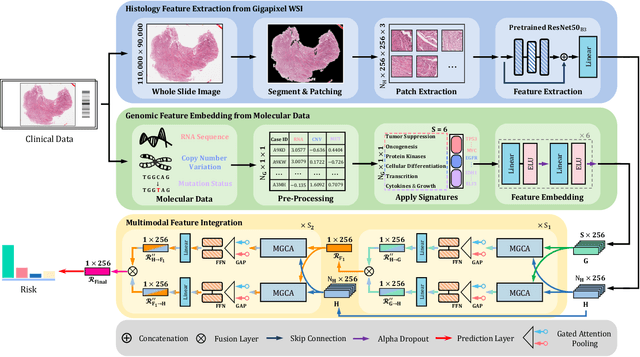
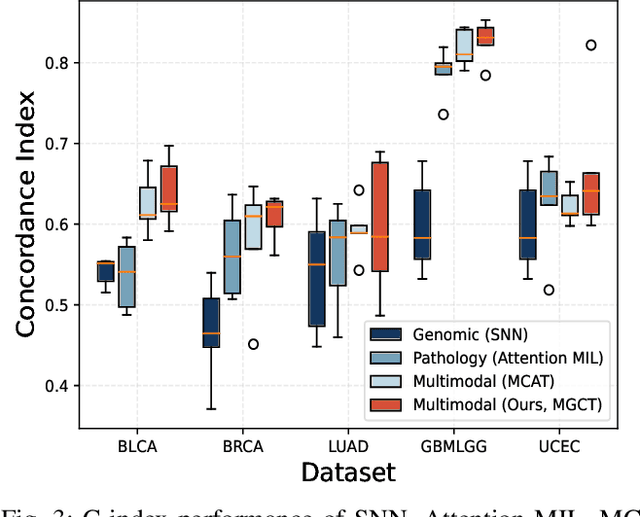
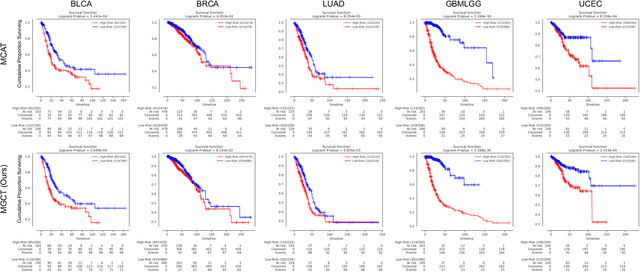
Abstract:The rapidly emerging field of deep learning-based computational pathology has shown promising results in utilizing whole slide images (WSIs) to objectively prognosticate cancer patients. However, most prognostic methods are currently limited to either histopathology or genomics alone, which inevitably reduces their potential to accurately predict patient prognosis. Whereas integrating WSIs and genomic features presents three main challenges: (1) the enormous heterogeneity of gigapixel WSIs which can reach sizes as large as 150,000x150,000 pixels; (2) the absence of a spatially corresponding relationship between histopathology images and genomic molecular data; and (3) the existing early, late, and intermediate multimodal feature fusion strategies struggle to capture the explicit interactions between WSIs and genomics. To ameliorate these issues, we propose the Mutual-Guided Cross-Modality Transformer (MGCT), a weakly-supervised, attention-based multimodal learning framework that can combine histology features and genomic features to model the genotype-phenotype interactions within the tumor microenvironment. To validate the effectiveness of MGCT, we conduct experiments using nearly 3,600 gigapixel WSIs across five different cancer types sourced from The Cancer Genome Atlas (TCGA). Extensive experimental results consistently emphasize that MGCT outperforms the state-of-the-art (SOTA) methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge