Guo-Wei Wei
Persistent Sheaf Laplacian Analysis of Protein Stability and Solubility Changes upon Mutation
Jan 18, 2026Abstract:Genetic mutations frequently disrupt protein structure, stability, and solubility, acting as primary drivers for a wide spectrum of diseases. Despite the critical importance of these molecular alterations, existing computational models often lack interpretability, and fail to integrate essential physicochemical interaction. To overcome these limitations, we propose SheafLapNet, a unified predictive framework grounded in the mathematical theory of Topological Deep Learning (TDL) and Persistent Sheaf Laplacian (PSL). Unlike standard Topological Data Analysis (TDA) tools such as persistent homology, which are often insensitive to heterogeneous information, PSL explicitly encodes specific physical and chemical information such as partial charges directly into the topological analysis. SheafLapNet synergizes these sheaf-theoretic invariants with advanced protein transformer features and auxiliary physical descriptors to capture intrinsic molecular interactions in a multiscale and mechanistic manner. To validate our framework, we employ rigorous benchmarks for both regression and classification tasks. For stability prediction, we utilize the comprehensive S2648 and S350 datasets. For solubility prediction, we employ the PON-Sol2 dataset, which provides annotations for increased, decreased, or neutral solubility changes. By integrating these multi-perspective features, SheafLapNet achieves state-of-the-art performance across these diverse benchmarks, demonstrating that sheaf-theoretic modeling significantly enhances both interpretability and generalizability in predicting mutation-induced structural and functional changes.
Multiscale Grassmann Manifolds for Single-Cell Data Analysis
Nov 12, 2025Abstract:Single-cell data analysis seeks to characterize cellular heterogeneity based on high-dimensional gene expression profiles. Conventional approaches represent each cell as a vector in Euclidean space, which limits their ability to capture intrinsic correlations and multiscale geometric structures. We propose a multiscale framework based on Grassmann manifolds that integrates machine learning with subspace geometry for single-cell data analysis. By generating embeddings under multiple representation scales, the framework combines their features from different geometric views into a unified Grassmann manifold. A power-based scale sampling function is introduced to control the selection of scales and balance in- formation across resolutions. Experiments on nine benchmark single-cell RNA-seq datasets demonstrate that the proposed approach effectively preserves meaningful structures and provides stable clustering performance, particularly for small to medium-sized datasets. These results suggest that Grassmann manifolds offer a coherent and informative foundation for analyzing single cell data.
Proteomic Learning of Gamma-Aminobutyric Acid (GABA) Receptor-Mediated Anesthesia
Jan 06, 2025



Abstract:Anesthetics are crucial in surgical procedures and therapeutic interventions, but they come with side effects and varying levels of effectiveness, calling for novel anesthetic agents that offer more precise and controllable effects. Targeting Gamma-aminobutyric acid (GABA) receptors, the primary inhibitory receptors in the central nervous system, could enhance their inhibitory action, potentially reducing side effects while improving the potency of anesthetics. In this study, we introduce a proteomic learning of GABA receptor-mediated anesthesia based on 24 GABA receptor subtypes by considering over 4000 proteins in protein-protein interaction (PPI) networks and over 1.5 millions known binding compounds. We develop a corresponding drug-target interaction network to identify potential lead compounds for novel anesthetic design. To ensure robust proteomic learning predictions, we curated a dataset comprising 136 targets from a pool of 980 targets within the PPI networks. We employed three machine learning algorithms, integrating advanced natural language processing (NLP) models such as pretrained transformer and autoencoder embeddings. Through a comprehensive screening process, we evaluated the side effects and repurposing potential of over 180,000 drug candidates targeting the GABRA5 receptor. Additionally, we assessed the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties of these candidates to identify those with near-optimal characteristics. This approach also involved optimizing the structures of existing anesthetics. Our work presents an innovative strategy for the development of new anesthetic drugs, optimization of anesthetic use, and deeper understanding of potential anesthesia-related side effects.
Persistent de Rham-Hodge Laplacians in the Eulerian representation
Aug 01, 2024Abstract:Recently, topological data analysis (TDA) has become a trending topic in data science and engineering. However, the key technique of TDA, i.e., persistent homology, is defined on point cloud data, which restricts its scope. In this work, we propose persistent de Rham-Hodge Laplacian, or persistent Hodge Laplacian (PHL) for abbreviation, for the TDA on manifolds with boundaries, or volumetric data. Specifically, we extended the evolutionary de Rham-Hodge theory from the Lagrangian formulation to the Eulerian formulation via structure-persevering Cartesian grids, and extended the persistent Laplacian on point clouds to persistent (de Rham-)Hodge Laplacian on nested families of manifolds with appropriate boundary conditions. The proposed PHL facilitates the machine learning and deep learning prediction of volumetric data. For a proof-of-principle application of the proposed PHL, we propose a persistent Hodge Laplacian learning (PHLL) algorithm for data on manifolds or volumetric data. To this end, we showcase the PHLL prediction of protein-ligand binding affinities in two benchmark datasets. Our numerical experiments highlight the power and promise of PHLL.
Graph-Based Bidirectional Transformer Decision Threshold Adjustment Algorithm for Class-Imbalanced Molecular Data
Jun 10, 2024
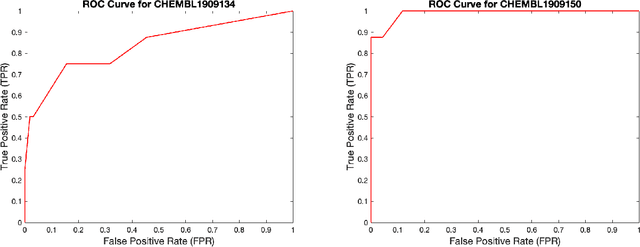
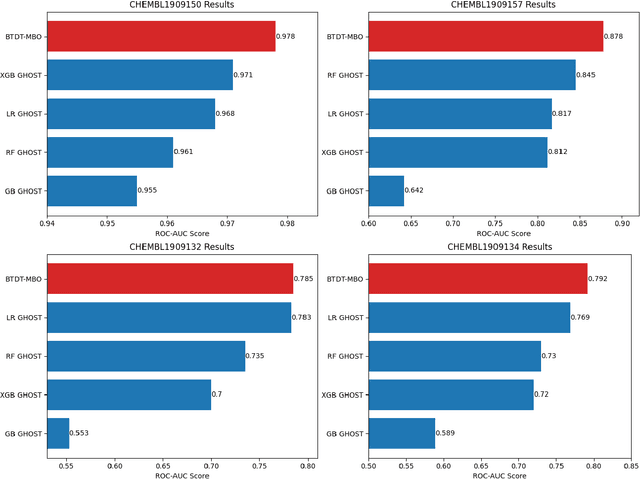
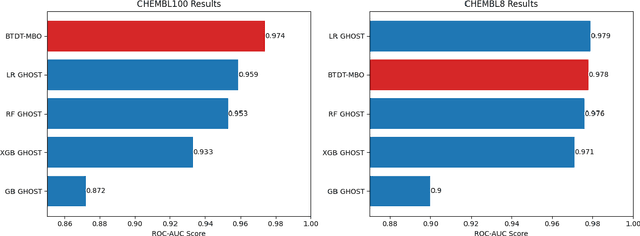
Abstract:Data sets with imbalanced class sizes, often where one class size is much smaller than that of others, occur extremely often in various applications, including those with biological foundations, such as drug discovery and disease diagnosis. Thus, it is extremely important to be able to identify data elements of classes of various sizes, as a failure to detect can result in heavy costs. However, many data classification algorithms do not perform well on imbalanced data sets as they often fail to detect elements belonging to underrepresented classes. In this paper, we propose the BTDT-MBO algorithm, incorporating Merriman-Bence-Osher (MBO) techniques and a bidirectional transformer, as well as distance correlation and decision threshold adjustments, for data classification problems on highly imbalanced molecular data sets, where the sizes of the classes vary greatly. The proposed method not only integrates adjustments in the classification threshold for the MBO algorithm in order to help deal with the class imbalance, but also uses a bidirectional transformer model based on an attention mechanism for self-supervised learning. Additionally, the method implements distance correlation as a weight function for the similarity graph-based framework on which the adjusted MBO algorithm operates. The proposed model is validated using six molecular data sets, and we also provide a thorough comparison to other competing algorithms. The computational experiments show that the proposed method performs better than competing techniques even when the class imbalance ratio is very high.
Position Paper: Challenges and Opportunities in Topological Deep Learning
Feb 14, 2024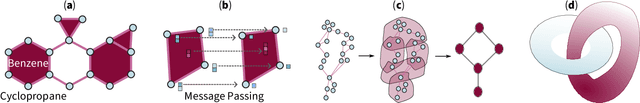
Abstract:Topological deep learning (TDL) is a rapidly evolving field that uses topological features to understand and design deep learning models. This paper posits that TDL may complement graph representation learning and geometric deep learning by incorporating topological concepts, and can thus provide a natural choice for various machine learning settings. To this end, this paper discusses open problems in TDL, ranging from practical benefits to theoretical foundations. For each problem, it outlines potential solutions and future research opportunities. At the same time, this paper serves as an invitation to the scientific community to actively participate in TDL research to unlock the potential of this emerging field.
Multiscale Topology in Interactomic Network: From Transcriptome to Antiaddiction Drug Repurposing
Dec 03, 2023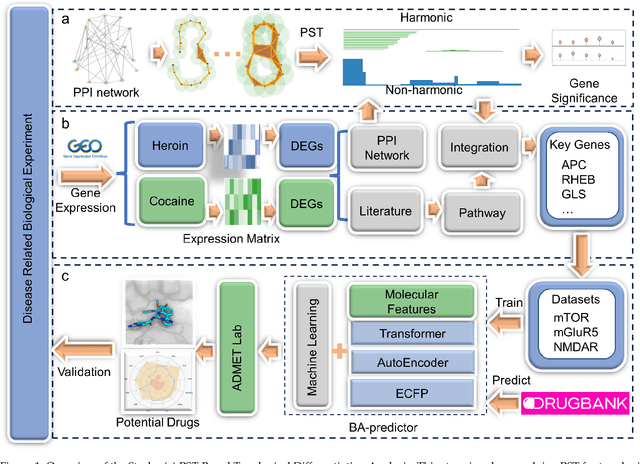
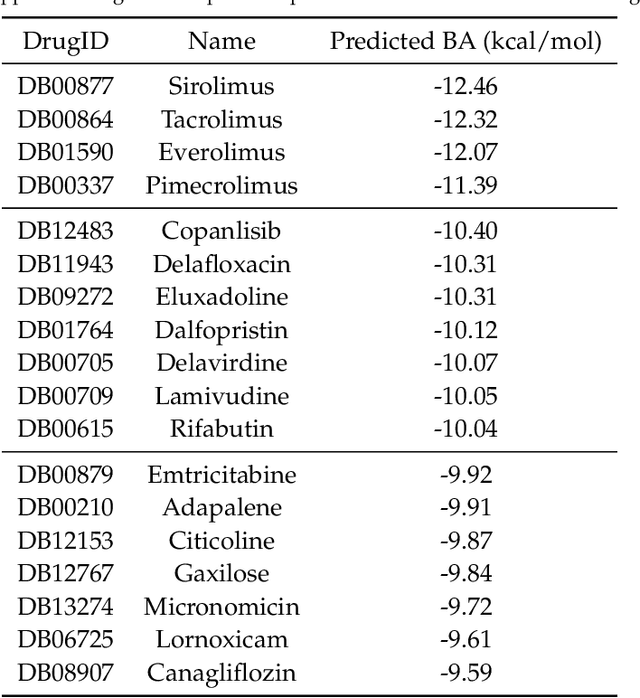
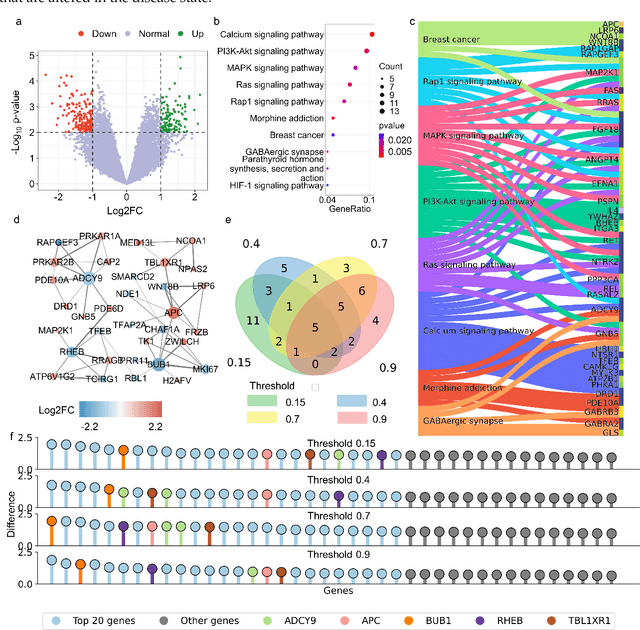
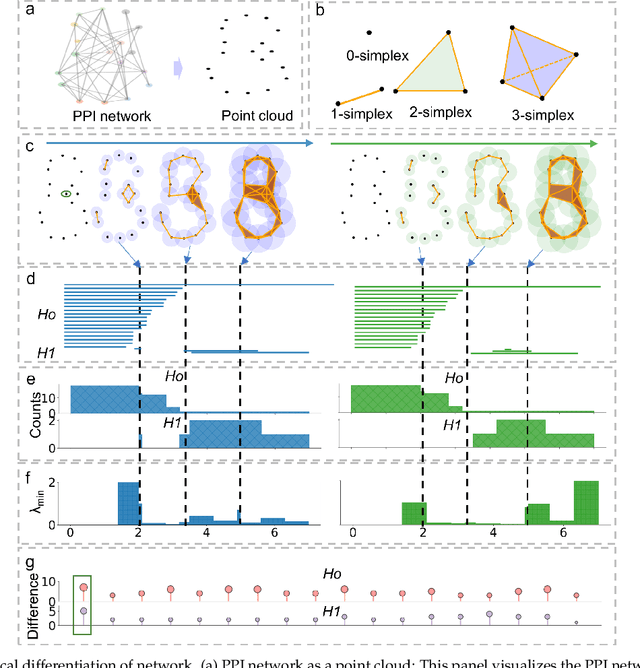
Abstract:The escalating drug addiction crisis in the United States underscores the urgent need for innovative therapeutic strategies. This study embarked on an innovative and rigorous strategy to unearth potential drug repurposing candidates for opioid and cocaine addiction treatment, bridging the gap between transcriptomic data analysis and drug discovery. We initiated our approach by conducting differential gene expression analysis on addiction-related transcriptomic data to identify key genes. We propose a novel topological differentiation to identify key genes from a protein-protein interaction (PPI) network derived from DEGs. This method utilizes persistent Laplacians to accurately single out pivotal nodes within the network, conducting this analysis in a multiscale manner to ensure high reliability. Through rigorous literature validation, pathway analysis, and data-availability scrutiny, we identified three pivotal molecular targets, mTOR, mGluR5, and NMDAR, for drug repurposing from DrugBank. We crafted machine learning models employing two natural language processing (NLP)-based embeddings and a traditional 2D fingerprint, which demonstrated robust predictive ability in gauging binding affinities of DrugBank compounds to selected targets. Furthermore, we elucidated the interactions of promising drugs with the targets and evaluated their drug-likeness. This study delineates a multi-faceted and comprehensive analytical framework, amalgamating bioinformatics, topological data analysis and machine learning, for drug repurposing in addiction treatment, setting the stage for subsequent experimental validation. The versatility of the methods we developed allows for applications across a range of diseases and transcriptomic datasets.
Analyzing Single Cell RNA Sequencing with Topological Nonnegative Matrix Factorization
Oct 24, 2023Abstract:Single-cell RNA sequencing (scRNA-seq) is a relatively new technology that has stimulated enormous interest in statistics, data science, and computational biology due to the high dimensionality, complexity, and large scale associated with scRNA-seq data. Nonnegative matrix factorization (NMF) offers a unique approach due to its meta-gene interpretation of resulting low-dimensional components. However, NMF approaches suffer from the lack of multiscale analysis. This work introduces two persistent Laplacian regularized NMF methods, namely, topological NMF (TNMF) and robust topological NMF (rTNMF). By employing a total of 12 datasets, we demonstrate that the proposed TNMF and rTNMF significantly outperform all other NMF-based methods. We have also utilized TNMF and rTNMF for the visualization of popular Uniform Manifold Approximation and Projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE).
K-Nearest-Neighbors Induced Topological PCA for scRNA Sequence Data Analysis
Oct 23, 2023



Abstract:Single-cell RNA sequencing (scRNA-seq) is widely used to reveal heterogeneity in cells, which has given us insights into cell-cell communication, cell differentiation, and differential gene expression. However, analyzing scRNA-seq data is a challenge due to sparsity and the large number of genes involved. Therefore, dimensionality reduction and feature selection are important for removing spurious signals and enhancing downstream analysis. Traditional PCA, a main workhorse in dimensionality reduction, lacks the ability to capture geometrical structure information embedded in the data, and previous graph Laplacian regularizations are limited by the analysis of only a single scale. We propose a topological Principal Components Analysis (tPCA) method by the combination of persistent Laplacian (PL) technique and L$_{2,1}$ norm regularization to address multiscale and multiclass heterogeneity issues in data. We further introduce a k-Nearest-Neighbor (kNN) persistent Laplacian technique to improve the robustness of our persistent Laplacian method. The proposed kNN-PL is a new algebraic topology technique which addresses the many limitations of the traditional persistent homology. Rather than inducing filtration via the varying of a distance threshold, we introduced kNN-tPCA, where filtrations are achieved by varying the number of neighbors in a kNN network at each step, and find that this framework has significant implications for hyper-parameter tuning. We validate the efficacy of our proposed tPCA and kNN-tPCA methods on 11 diverse benchmark scRNA-seq datasets, and showcase that our methods outperform other unsupervised PCA enhancements from the literature, as well as popular Uniform Manifold Approximation (UMAP), t-Distributed Stochastic Neighbor Embedding (tSNE), and Projection Non-Negative Matrix Factorization (NMF) by significant margins.
ChatGPT for Computational Topology
Oct 19, 2023Abstract:ChatGPT represents a significant milestone in the field of artificial intelligence (AI), finding widespread applications across diverse domains. However, its effectiveness in mathematical contexts has been somewhat constrained by its susceptibility to conceptual errors. Concurrently, topological data analysis (TDA), a relatively new discipline, has garnered substantial interest in recent years. Nonetheless, the advancement of TDA is impeded by the limited understanding of computational algorithms and coding proficiency among theoreticians. This work endeavors to bridge the gap between theoretical topological concepts and their practical implementation in computational topology through the utilization of ChatGPT. We showcase how a pure theoretician, devoid of computational experience and coding skills, can effectively transform mathematical formulations and concepts into functional code for computational topology with the assistance of ChatGPT. Our strategy outlines a productive process wherein a mathematician trains ChatGPT on pure mathematical concepts, steers ChatGPT towards generating computational topology code, and subsequently validates the generated code using established examples. Our specific case studies encompass the computation of Betti numbers, Laplacian matrices, and Dirac matrices for simplicial complexes, as well as the persistence of various homologies and Laplacians. Furthermore, we explore the application of ChatGPT in computing recently developed topological theories for hypergraphs and digraphs. This work serves as an initial step towards effectively transforming pure mathematical theories into practical computational tools, with the ultimate goal of enabling real applications across diverse fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge