Ganapathy Krishnamurthi
Exact Constraint Enforcement in Physics-Informed Extreme Learning Machines using Null-Space Projection Framework
Jan 16, 2026Abstract:Physics-informed extreme learning machines (PIELMs) typically impose boundary and initial conditions through penalty terms, yielding only approximate satisfaction that is sensitive to user-specified weights and can propagate errors into the interior solution. This work introduces Null-Space Projected PIELM (NP-PIELM), achieving exact constraint enforcement through algebraic projection in coefficient space. The method exploits the geometric structure of the admissible coefficient manifold, recognizing that it admits a decomposition through the null space of the boundary operator. By characterizing this manifold via a translation-invariant representation and projecting onto the kernel component, optimization is restricted to constraint-preserving directions, transforming the constrained problem into unconstrained least-squares where boundary conditions are satisfied exactly at discrete collocation points. This eliminates penalty coefficients, dual variables, and problem-specific constructions while preserving single-shot training efficiency. Numerical experiments on elliptic and parabolic problems including complex geometries and mixed boundary conditions validate the framework.
MedPAO: A Protocol-Driven Agent for Structuring Medical Reports
Oct 06, 2025Abstract:The deployment of Large Language Models (LLMs) for structuring clinical data is critically hindered by their tendency to hallucinate facts and their inability to follow domain-specific rules. To address this, we introduce MedPAO, a novel agentic framework that ensures accuracy and verifiable reasoning by grounding its operation in established clinical protocols such as the ABCDEF protocol for CXR analysis. MedPAO decomposes the report structuring task into a transparent process managed by a Plan-Act-Observe (PAO) loop and specialized tools. This protocol-driven method provides a verifiable alternative to opaque, monolithic models. The efficacy of our approach is demonstrated through rigorous evaluation: MedPAO achieves an F1-score of 0.96 on the critical sub-task of concept categorization. Notably, expert radiologists and clinicians rated the final structured outputs with an average score of 4.52 out of 5, indicating a level of reliability that surpasses baseline approaches relying solely on LLM-based foundation models. The code is available at: https://github.com/MiRL-IITM/medpao-agent
* Paper published at "Agentic AI for Medicine" Workshop, MICCAI 2025
Efficient Knowledge Distillation of SAM for Medical Image Segmentation
Jan 28, 2025



Abstract:The Segment Anything Model (SAM) has set a new standard in interactive image segmentation, offering robust performance across various tasks. However, its significant computational requirements limit its deployment in real-time or resource-constrained environments. To address these challenges, we propose a novel knowledge distillation approach, KD SAM, which incorporates both encoder and decoder optimization through a combination of Mean Squared Error (MSE) and Perceptual Loss. This dual-loss framework captures structural and semantic features, enabling the student model to maintain high segmentation accuracy while reducing computational complexity. Based on the model evaluation on datasets, including Kvasir-SEG, ISIC 2017, Fetal Head Ultrasound, and Breast Ultrasound, we demonstrate that KD SAM achieves comparable or superior performance to the baseline models, with significantly fewer parameters. KD SAM effectively balances segmentation accuracy and computational efficiency, making it well-suited for real-time medical image segmentation applications in resource-constrained environments.
PICS in Pics: Physics Informed Contour Selection for Rapid Image Segmentation
Nov 13, 2023Abstract:Effective training of deep image segmentation models is challenging due to the need for abundant, high-quality annotations. Generating annotations is laborious and time-consuming for human experts, especially in medical image segmentation. To facilitate image annotation, we introduce Physics Informed Contour Selection (PICS) - an interpretable, physics-informed algorithm for rapid image segmentation without relying on labeled data. PICS draws inspiration from physics-informed neural networks (PINNs) and an active contour model called snake. It is fast and computationally lightweight because it employs cubic splines instead of a deep neural network as a basis function. Its training parameters are physically interpretable because they directly represent control knots of the segmentation curve. Traditional snakes involve minimization of the edge-based loss functionals by deriving the Euler-Lagrange equation followed by its numerical solution. However, PICS directly minimizes the loss functional, bypassing the Euler Lagrange equations. It is the first snake variant to minimize a region-based loss function instead of traditional edge-based loss functions. PICS uniquely models the three-dimensional (3D) segmentation process with an unsteady partial differential equation (PDE), which allows accelerated segmentation via transfer learning. To demonstrate its effectiveness, we apply PICS for 3D segmentation of the left ventricle on a publicly available cardiac dataset. While doing so, we also introduce a new convexity-preserving loss term that encodes the shape information of the left ventricle to enhance PICS's segmentation quality. Overall, PICS presents several novelties in network architecture, transfer learning, and physics-inspired losses for image segmentation, thereby showing promising outcomes and potential for further refinement.
PARSE challenge 2022: Pulmonary Arteries Segmentation using Swin U-Net Transformer(Swin UNETR) and U-Net
Aug 20, 2022

Abstract:In this work, we present our proposed method to segment the pulmonary arteries from the CT scans using Swin UNETR and U-Net-based deep neural network architecture. Six models, three models based on Swin UNETR, and three models based on 3D U-net with residual units were ensemble using a weighted average to make the final segmentation masks. Our team achieved a multi-level dice score of 84.36 percent through this method. The code of our work is available on the following link: https://github.com/akansh12/parse2022. This work is part of the MICCAI PARSE 2022 challenge.
Brain Tumor Segmentation and Survival Prediction using Automatic Hard mining in 3D CNN Architecture
Jan 05, 2021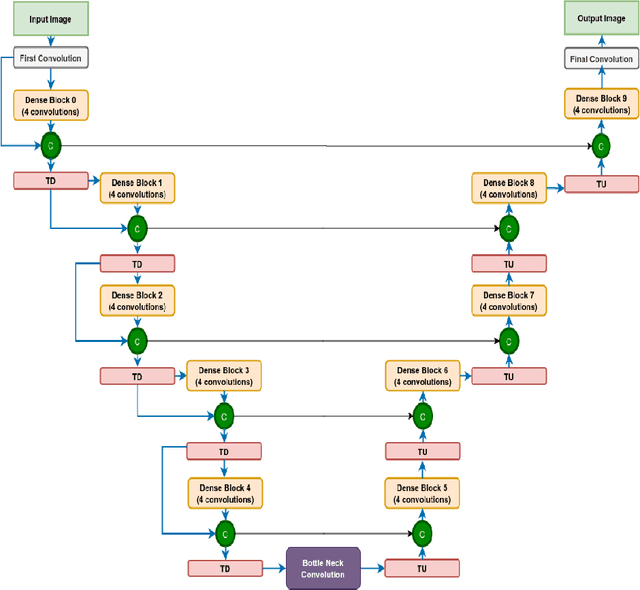
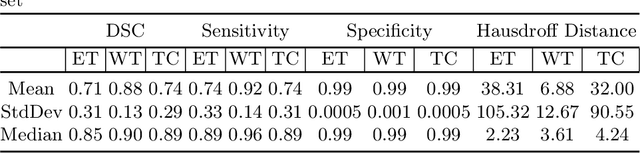
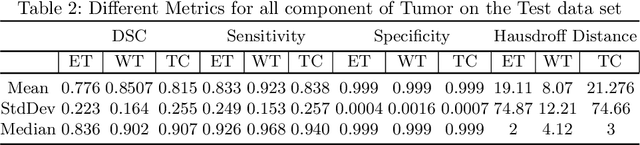
Abstract:We utilize 3-D fully convolutional neural networks (CNN) to segment gliomas and its constituents from multimodal Magnetic Resonance Images (MRI). The architecture uses dense connectivity patterns to reduce the number of weights and residual connections and is initialized with weights obtained from training this model with BraTS 2018 dataset. Hard mining is done during training to train for the difficult cases of segmentation tasks by increasing the dice similarity coefficient (DSC) threshold to choose the hard cases as epoch increases. On the BraTS2020 validation data (n = 125), this architecture achieved a tumor core, whole tumor, and active tumor dice of 0.744, 0.876, 0.714,respectively. On the test dataset, we get an increment in DSC of tumor core and active tumor by approximately 7%. In terms of DSC, our network performances on the BraTS 2020 test data are 0.775, 0.815, and 0.85 for enhancing tumor, tumor core, and whole tumor, respectively. Overall survival of a subject is determined using conventional machine learning from rediomics features obtained using a generated segmentation mask. Our approach has achieved 0.448 and 0.452 as the accuracy on the validation and test dataset.
Abstracting Deep Neural Networks into Concept Graphs for Concept Level Interpretability
Aug 14, 2020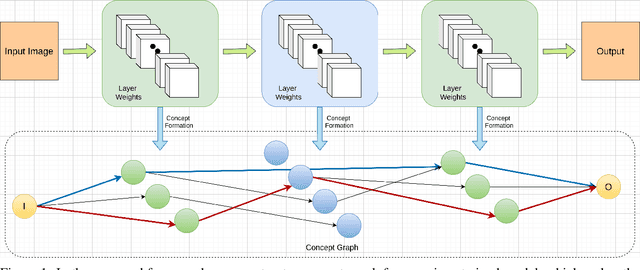

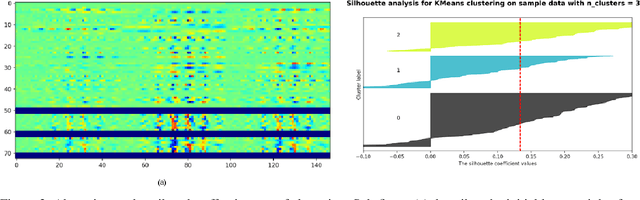
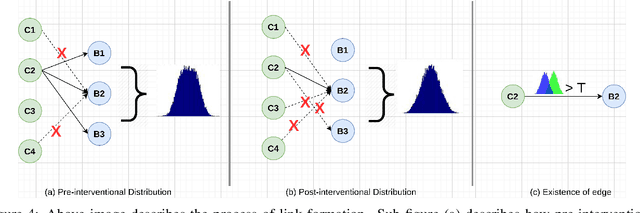
Abstract:The black-box nature of deep learning models prevents them from being completely trusted in domains like biomedicine. Most explainability techniques do not capture the concept-based reasoning that human beings follow. In this work, we attempt to understand the behavior of trained models that perform image processing tasks in the medical domain by building a graphical representation of the concepts they learn. Extracting such a graphical representation of the model's behavior on an abstract, higher conceptual level would unravel the learnings of these models and would help us to evaluate the steps taken by the model for predictions. We show the application of our proposed implementation on two biomedical problems - brain tumor segmentation and fundus image classification. We provide an alternative graphical representation of the model by formulating a \textit{concept level graph} as discussed above, which makes the problem of intervention to find active inference trails more tractable. Understanding these trails would provide an understanding of the hierarchy of the decision-making process followed by the model. [As well as overall nature of model]. Our framework is available at \url{https://github.com/koriavinash1/BioExp}
Distangling Biological Noise in Cellular Images with a focus on Explainability
Jul 11, 2020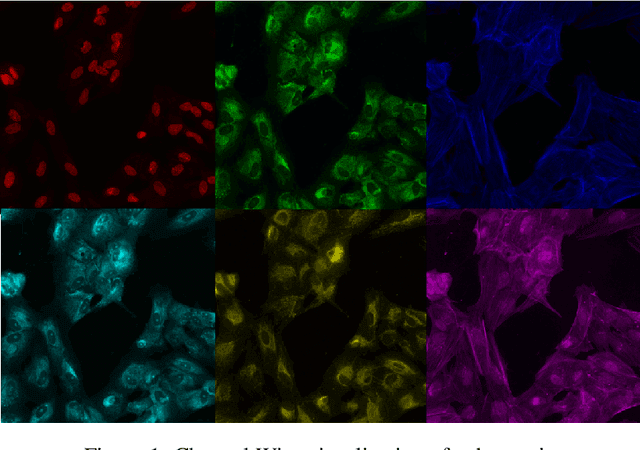
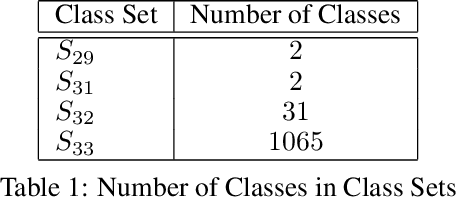
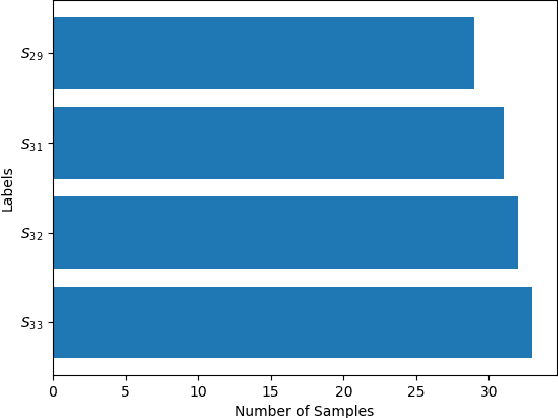
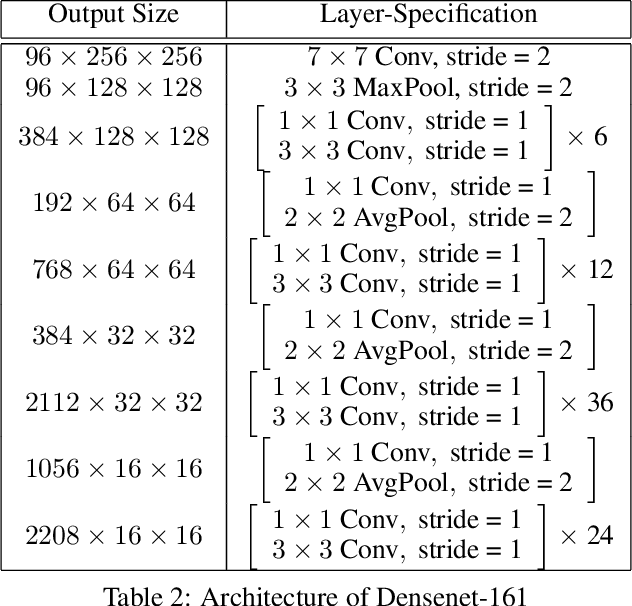
Abstract:The cost of some drugs and medical treatments has risen in recent years that many patients are having to go without. A classification project could make researchers more efficient. One of the more surprising reasons behind the cost is how long it takes to bring new treatments to market. Despite improvements in technology and science, research and development continues to lag. In fact, finding new treatment takes, on average, more than 10 years and costs hundreds of millions of dollars. In turn, greatly decreasing the cost of treatments can make ensure these treatments get to patients faster. This work aims at solving a part of this problem by creating a cellular image classification model which can decipher the genetic perturbations in cell (occurring naturally or artificially). Another interesting question addressed is what makes the deep-learning model decide in a particular fashion, which can further help in demystifying the mechanism of action of certain perturbations and paves a way towards the explainability of the deep-learning model. We show the results of Grad-CAM visualizations and make a case for the significance of certain features over others. Further we discuss how these significant features are pivotal in extracting useful diagnostic information from the deep-learning model.
Structurally aware bidirectional unpaired image to image translation between CT and MR
Jun 05, 2020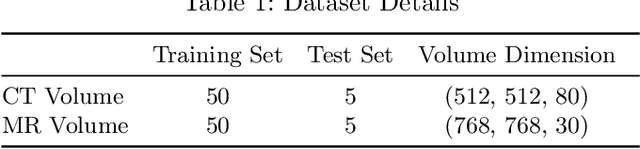
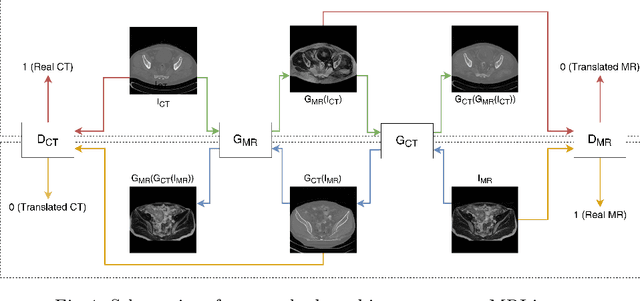
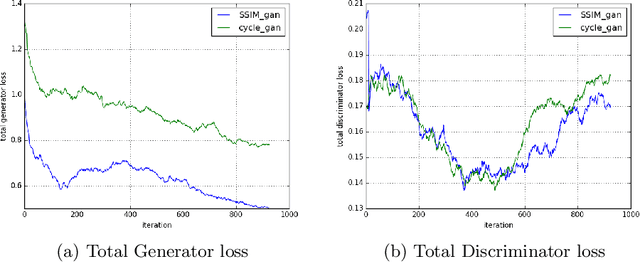
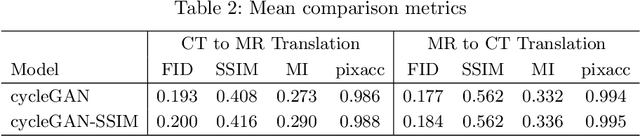
Abstract:Magnetic Resonance (MR) Imaging and Computed Tomography (CT) are the primary diagnostic imaging modalities quite frequently used for surgical planning and analysis. A general problem with medical imaging is that the acquisition process is quite expensive and time-consuming. Deep learning techniques like generative adversarial networks (GANs) can help us to leverage the possibility of an image to image translation between multiple imaging modalities, which in turn helps in saving time and cost. These techniques will help to conduct surgical planning under CT with the feedback of MRI information. While previous studies have shown paired and unpaired image synthesis from MR to CT, image synthesis from CT to MR still remains a challenge, since it involves the addition of extra tissue information. In this manuscript, we have implemented two different variations of Generative Adversarial Networks exploiting the cycling consistency and structural similarity between both CT and MR image modalities on a pelvis dataset, thus facilitating a bidirectional exchange of content and style between these image modalities. The proposed GANs translate the input medical images by different mechanisms, and hence generated images not only appears realistic but also performs well across various comparison metrics, and these images have also been cross verified with a radiologist. The radiologist verification has shown that slight variations in generated MR and CT images may not be exactly the same as their true counterpart but it can be used for medical purposes.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020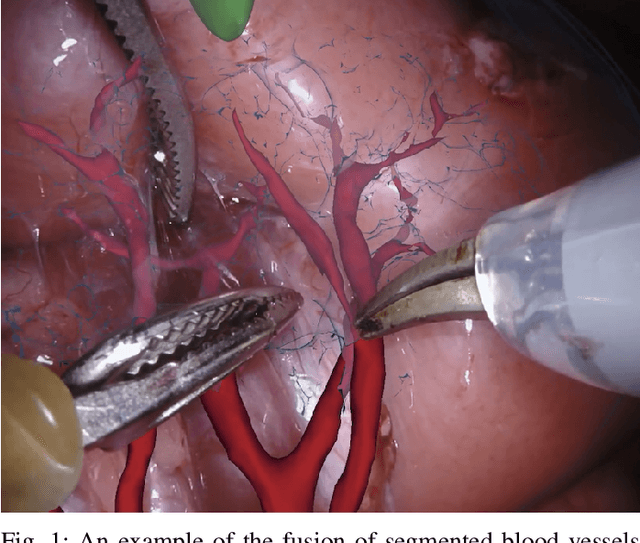
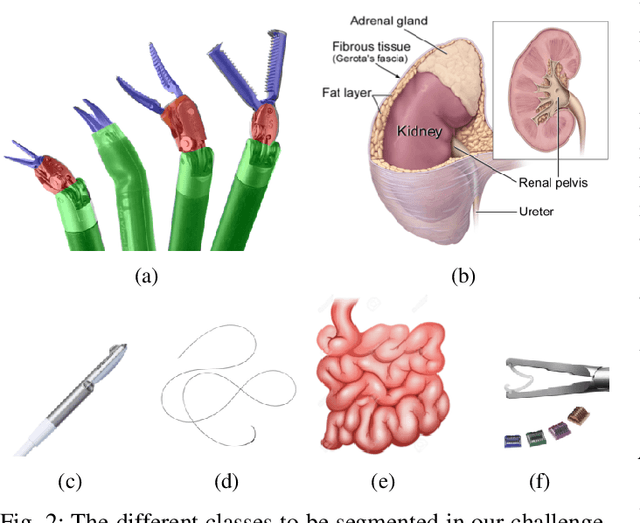
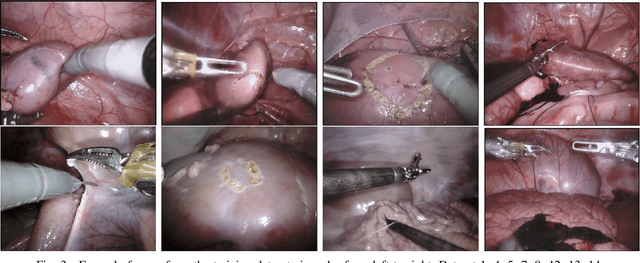
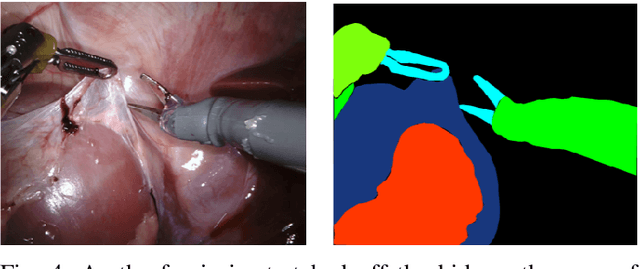
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge