Demystifying Brain Tumour Segmentation Networks: Interpretability and Uncertainty Analysis
Paper and Code
Sep 05, 2019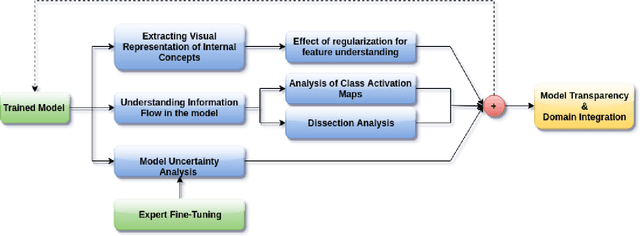
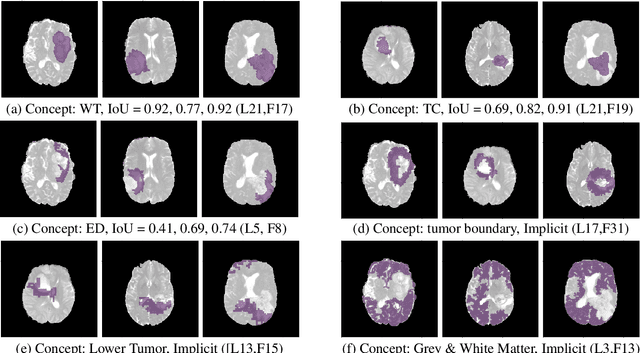
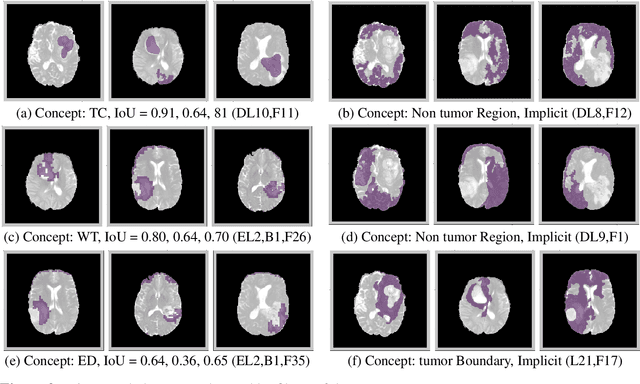
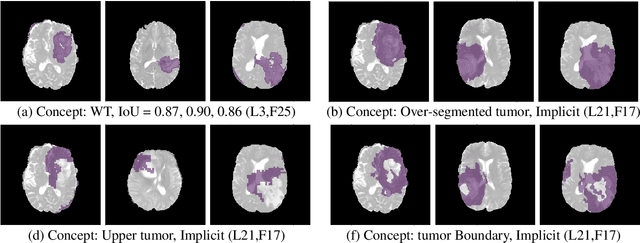
The accurate automatic segmentation of gliomas and its intra-tumoral structures is important not only for treatment planning but also for follow-up evaluations. Several methods based on 2D and 3D Deep Neural Networks (DNN) have been developed to segment brain tumors and to classify different categories of tumors from different MRI modalities. However, these networks are often black-box models and do not provide any evidence regarding the process they take to perform this task. Increasing transparency and interpretability of such deep learning techniques are necessary for the complete integration of such methods into medical practice. In this paper, we explore various techniques to explain the functional organization of brain tumor segmentation models and to extract visualizations of internal concepts to understand how these networks achieve highly accurate tumor segmentations. We use the BraTS 2018 dataset to train three different networks with standard architectures and outline similarities and differences in the process that these networks take to segment brain tumors. We show that brain tumor segmentation networks learn certain human-understandable disentangled concepts on a filter level. We also show that they take a top-down or hierarchical approach to localizing the different parts of the tumor. We then extract visualizations of some internal feature maps and also provide a measure of uncertainty with regards to the outputs of the models to give additional qualitative evidence about the predictions of these networks. We believe that the emergence of such human-understandable organization and concepts might aid in the acceptance and integration of such methods in medical diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge