Vikas Kumar Anand
Brain Tumor Segmentation and Survival Prediction using Automatic Hard mining in 3D CNN Architecture
Jan 05, 2021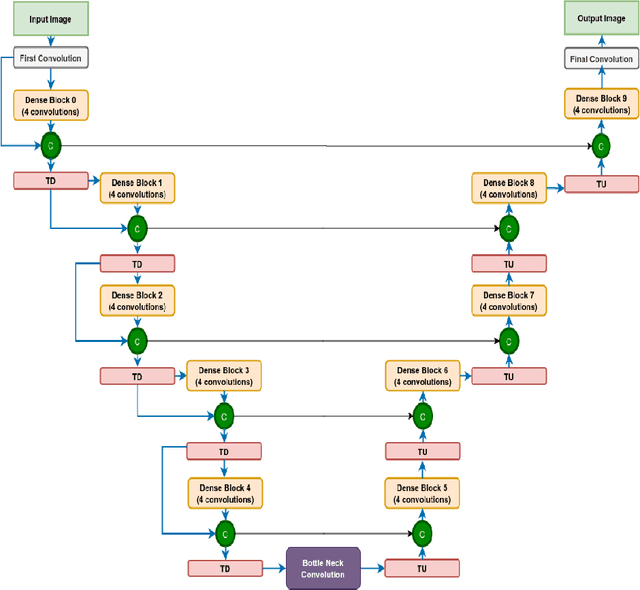
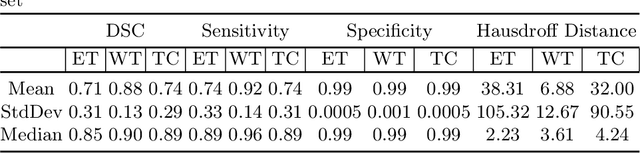
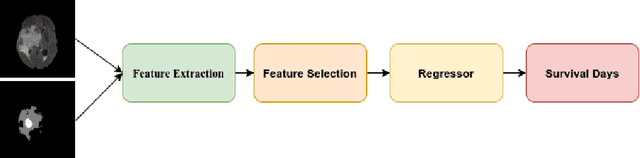
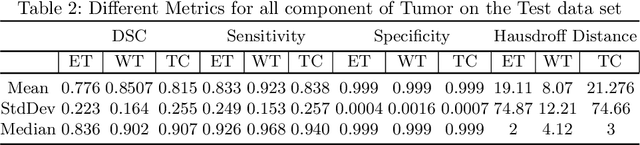
Abstract:We utilize 3-D fully convolutional neural networks (CNN) to segment gliomas and its constituents from multimodal Magnetic Resonance Images (MRI). The architecture uses dense connectivity patterns to reduce the number of weights and residual connections and is initialized with weights obtained from training this model with BraTS 2018 dataset. Hard mining is done during training to train for the difficult cases of segmentation tasks by increasing the dice similarity coefficient (DSC) threshold to choose the hard cases as epoch increases. On the BraTS2020 validation data (n = 125), this architecture achieved a tumor core, whole tumor, and active tumor dice of 0.744, 0.876, 0.714,respectively. On the test dataset, we get an increment in DSC of tumor core and active tumor by approximately 7%. In terms of DSC, our network performances on the BraTS 2020 test data are 0.775, 0.815, and 0.85 for enhancing tumor, tumor core, and whole tumor, respectively. Overall survival of a subject is determined using conventional machine learning from rediomics features obtained using a generated segmentation mask. Our approach has achieved 0.448 and 0.452 as the accuracy on the validation and test dataset.
Structurally aware bidirectional unpaired image to image translation between CT and MR
Jun 05, 2020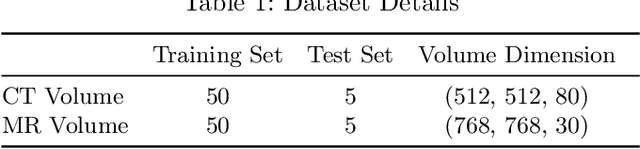
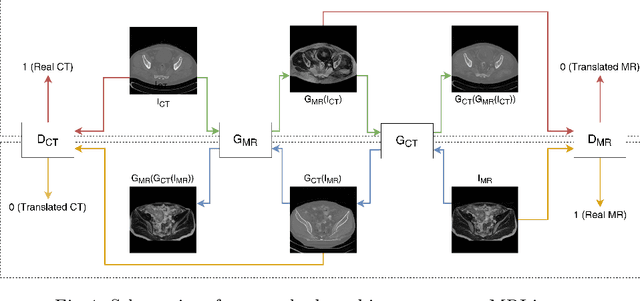
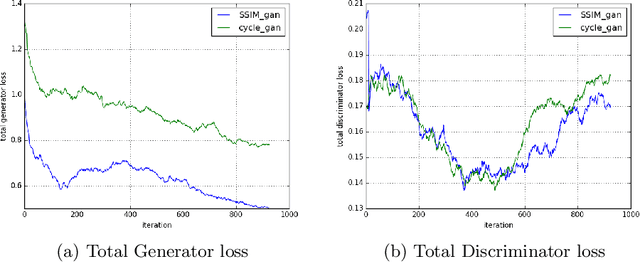
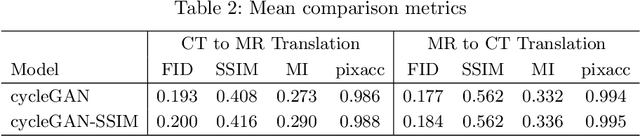
Abstract:Magnetic Resonance (MR) Imaging and Computed Tomography (CT) are the primary diagnostic imaging modalities quite frequently used for surgical planning and analysis. A general problem with medical imaging is that the acquisition process is quite expensive and time-consuming. Deep learning techniques like generative adversarial networks (GANs) can help us to leverage the possibility of an image to image translation between multiple imaging modalities, which in turn helps in saving time and cost. These techniques will help to conduct surgical planning under CT with the feedback of MRI information. While previous studies have shown paired and unpaired image synthesis from MR to CT, image synthesis from CT to MR still remains a challenge, since it involves the addition of extra tissue information. In this manuscript, we have implemented two different variations of Generative Adversarial Networks exploiting the cycling consistency and structural similarity between both CT and MR image modalities on a pelvis dataset, thus facilitating a bidirectional exchange of content and style between these image modalities. The proposed GANs translate the input medical images by different mechanisms, and hence generated images not only appears realistic but also performs well across various comparison metrics, and these images have also been cross verified with a radiologist. The radiologist verification has shown that slight variations in generated MR and CT images may not be exactly the same as their true counterpart but it can be used for medical purposes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge