Mahendra Khened
Brain Tumor Segmentation and Survival Prediction using Automatic Hard mining in 3D CNN Architecture
Jan 05, 2021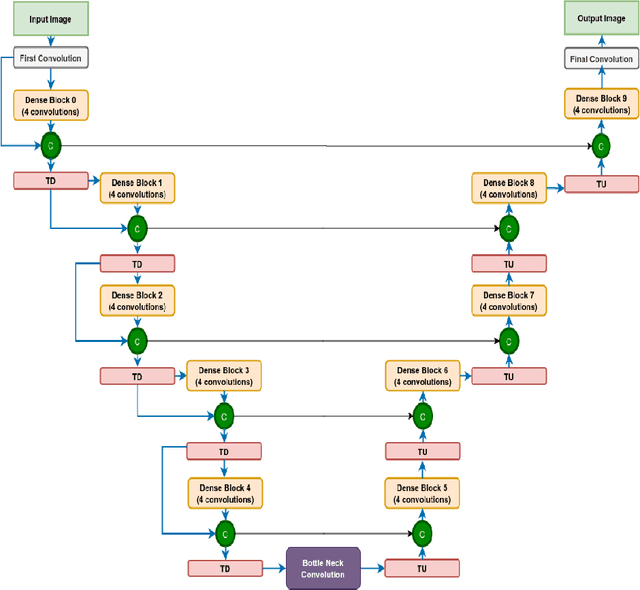
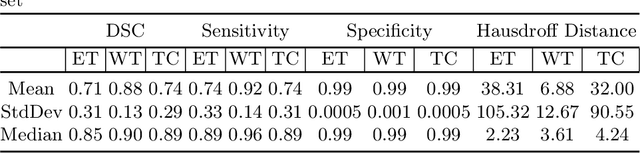
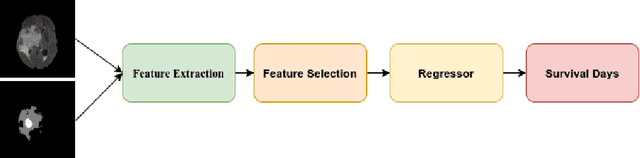
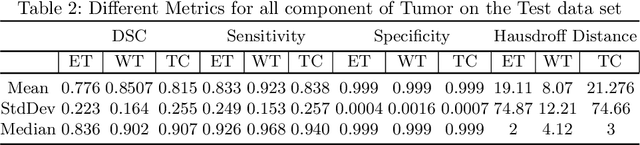
Abstract:We utilize 3-D fully convolutional neural networks (CNN) to segment gliomas and its constituents from multimodal Magnetic Resonance Images (MRI). The architecture uses dense connectivity patterns to reduce the number of weights and residual connections and is initialized with weights obtained from training this model with BraTS 2018 dataset. Hard mining is done during training to train for the difficult cases of segmentation tasks by increasing the dice similarity coefficient (DSC) threshold to choose the hard cases as epoch increases. On the BraTS2020 validation data (n = 125), this architecture achieved a tumor core, whole tumor, and active tumor dice of 0.744, 0.876, 0.714,respectively. On the test dataset, we get an increment in DSC of tumor core and active tumor by approximately 7%. In terms of DSC, our network performances on the BraTS 2020 test data are 0.775, 0.815, and 0.85 for enhancing tumor, tumor core, and whole tumor, respectively. Overall survival of a subject is determined using conventional machine learning from rediomics features obtained using a generated segmentation mask. Our approach has achieved 0.448 and 0.452 as the accuracy on the validation and test dataset.
A Generalized Deep Learning Framework for Whole-Slide Image Segmentation and Analysis
Jan 01, 2020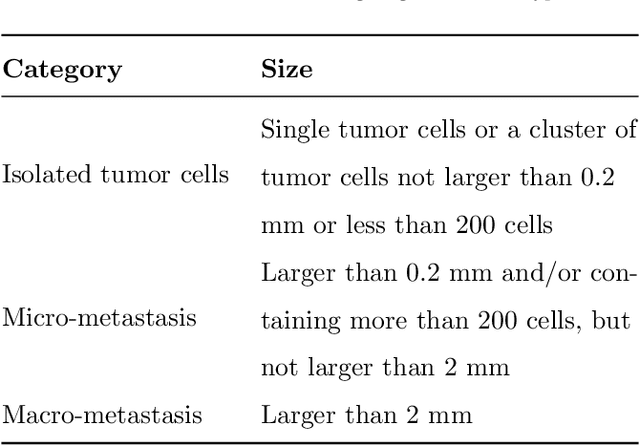
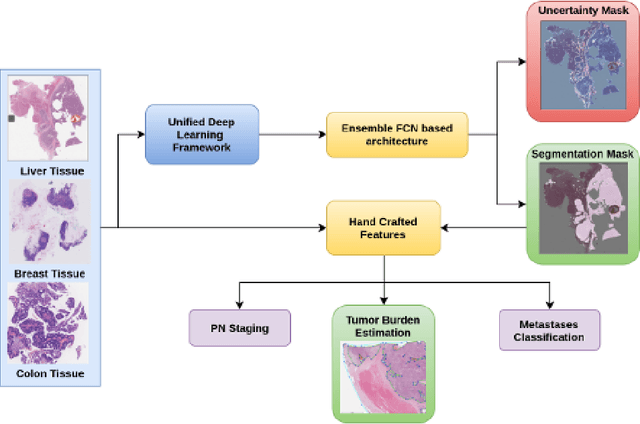
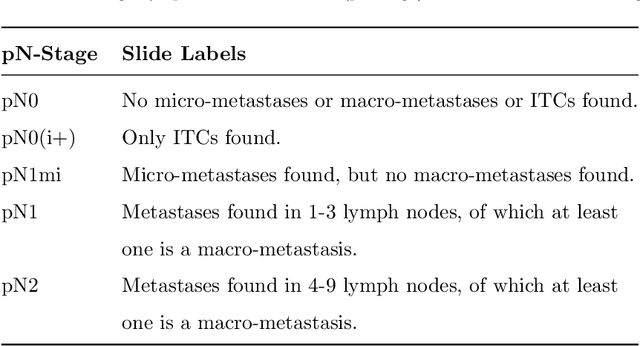
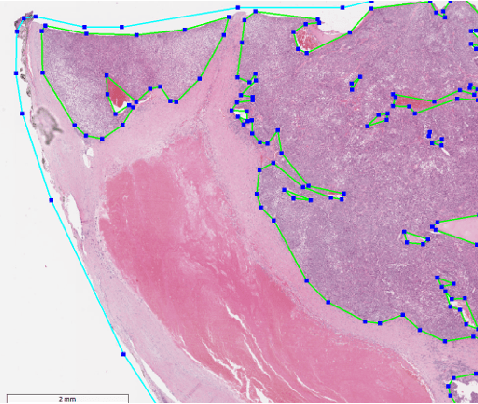
Abstract:Histopathology tissue analysis is considered the gold standard in cancer diagnosis and prognosis. Given the large size of these images and the increase in the number of potential cancer cases, an automated solution as an aid to histopathologists is highly desirable. In the recent past, deep learning-based techniques have provided state of the art results in a wide variety of image analysis tasks, including analysis of digitized slides. However, the size of images and variability in histopathology tasks makes it a challenge to develop an integrated framework for histopathology image analysis. We propose a deep learning-based framework for histopathology tissue analysis. We demonstrate the generalizability of our framework, including training and inference, on several open-source datasets, which include CAMELYON (breast cancer metastases), DigestPath (colon cancer), and PAIP (liver cancer) datasets. We discuss multiple types of uncertainties pertaining to data and model, namely aleatoric and epistemic, respectively. Simultaneously, we demonstrate our model generalization across different data distribution by evaluating some samples on TCGA data. On CAMELYON16 test data (n=139) for the task of lesion detection, the FROC score achieved was 0.86 and in the CAMELYON17 test-data (n=500) for the task of pN-staging the Cohen's kappa score achieved was 0.9090 (third in the open leaderboard). On DigestPath test data (n=212) for the task of tumor segmentation, a Dice score of 0.782 was achieved (fourth in the challenge). On PAIP test data (n=40) for the task of viable tumor segmentation, a Jaccard Index of 0.75 (third in the challenge) was achieved, and for viable tumor burden, a score of 0.633 was achieved (second in the challenge). Our entire framework and related documentation are freely available at GitHub and PyPi.
Fully Convolutional Multi-scale Residual DenseNets for Cardiac Segmentation and Automated Cardiac Diagnosis using Ensemble of Classifiers
Jan 16, 2018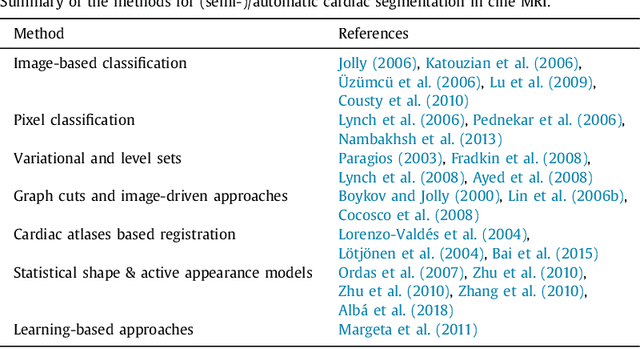
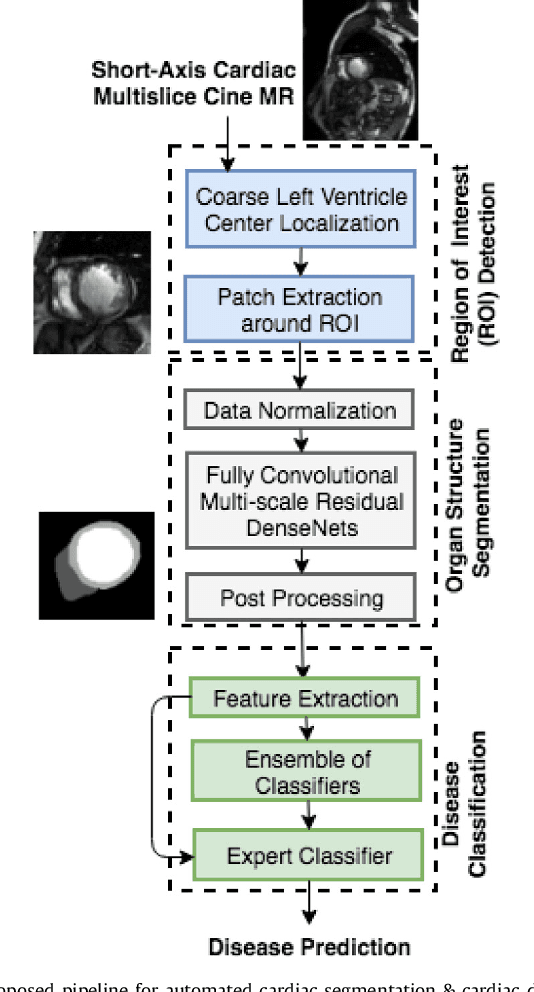
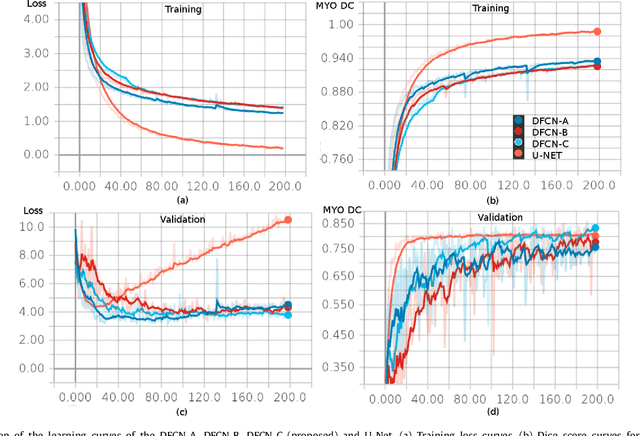

Abstract:Deep fully convolutional neural network (FCN) based architectures have shown great potential in medical image segmentation. However, such architectures usually have millions of parameters and inadequate number of training samples leading to over-fitting and poor generalization. In this paper, we present a novel highly parameter and memory efficient FCN based architecture for medical image analysis. We propose a novel up-sampling path which incorporates long skip and short-cut connections to overcome the feature map explosion in FCN like architectures. In order to processes the input images at multiple scales and view points simultaneously, we propose to incorporate Inception module's parallel structures. We also propose a novel dual loss function whose weighting scheme allows to combine advantages of cross-entropy and dice loss. We have validated our proposed network architecture on two publicly available datasets, namely: (i) Automated Cardiac Disease Diagnosis Challenge (ACDC-2017), (ii) Left Ventricular Segmentation Challenge (LV-2011). Our approach in ACDC-2017 challenge stands second place for segmentation and first place in automated cardiac disease diagnosis tasks with an accuracy of 100%. In the LV-2011 challenge our approach attained 0.74 Jaccard index, which is so far the highest published result in fully automated algorithms. From the segmentation we extracted clinically relevant cardiac parameters and hand-crafted features which reflected the clinical diagnostic analysis to train an ensemble system for cardiac disease classification. Our approach combined both cardiac segmentation and disease diagnosis into a fully automated framework which is computational efficient and hence has the potential to be incorporated in computer-aided diagnosis (CAD) tools for clinical application.
2D-Densely Connected Convolution Neural Networks for automatic Liver and Tumor Segmentation
Jan 05, 2018



Abstract:In this paper we propose a fully automatic 2-stage cascaded approach for segmentation of liver and its tumors in CT (Computed Tomography) images using densely connected fully convolutional neural network (DenseNet). We independently train liver and tumor segmentation models and cascade them for a combined segmentation of the liver and its tumor. The first stage involves segmentation of liver and the second stage uses the first stage's segmentation results for localization of liver and henceforth tumor segmentations inside liver region. The liver model was trained on the down-sampled axial slices $(256 \times 256)$, whereas for the tumor model no down-sampling of slices was done, but instead it was trained on the CT axial slices windowed at three different Hounsfield (HU) levels. On the test set our model achieved a global dice score of 0.923 and 0.625 on liver and tumor respectively. The computed tumor burden had an rmse of 0.044.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge