Balaji Srinivasan
Exact Constraint Enforcement in Physics-Informed Extreme Learning Machines using Null-Space Projection Framework
Jan 16, 2026Abstract:Physics-informed extreme learning machines (PIELMs) typically impose boundary and initial conditions through penalty terms, yielding only approximate satisfaction that is sensitive to user-specified weights and can propagate errors into the interior solution. This work introduces Null-Space Projected PIELM (NP-PIELM), achieving exact constraint enforcement through algebraic projection in coefficient space. The method exploits the geometric structure of the admissible coefficient manifold, recognizing that it admits a decomposition through the null space of the boundary operator. By characterizing this manifold via a translation-invariant representation and projecting onto the kernel component, optimization is restricted to constraint-preserving directions, transforming the constrained problem into unconstrained least-squares where boundary conditions are satisfied exactly at discrete collocation points. This eliminates penalty coefficients, dual variables, and problem-specific constructions while preserving single-shot training efficiency. Numerical experiments on elliptic and parabolic problems including complex geometries and mixed boundary conditions validate the framework.
Deep vs. Shallow: Benchmarking Physics-Informed Neural Architectures on the Biharmonic Equation
Oct 06, 2025Abstract:Partial differential equation (PDE) solvers are fundamental to engineering simulation. Classical mesh-based approaches (finite difference/volume/element) are fast and accurate on high-quality meshes but struggle with higher-order operators and complex, hard-to-mesh geometries. Recently developed physics-informed neural networks (PINNs) and their variants are mesh-free and flexible, yet compute-intensive and often less accurate. This paper systematically benchmarks RBF-PIELM, a rapid PINN variant-an extreme learning machine with radial-basis activations-for higher-order PDEs. RBF-PIELM replaces PINNs' time-consuming gradient descent with a single-shot least-squares solve. We test RBF-PIELM on the fourth-order biharmonic equation using two benchmarks: lid-driven cavity flow (streamfunction formulation) and a manufactured oscillatory solution. Our results show up to $(350\times)$ faster training than PINNs and over $(10\times)$ fewer parameters for comparable solution accuracy. Despite surpassing PINNs, RBF-PIELM still lags mature mesh-based solvers and its accuracy degrades on highly oscillatory solutions, highlighting remaining challenges for practical deployment.
Towards Fast Option Pricing PDE Solvers Powered by PIELM
Oct 05, 2025Abstract:Partial differential equation (PDE) solvers underpin modern quantitative finance, governing option pricing and risk evaluation. Physics-Informed Neural Networks (PINNs) have emerged as a promising approach for solving the forward and inverse problems of partial differential equations (PDEs) using deep learning. However they remain computationally expensive due to their iterative gradient descent based optimization and scale poorly with increasing model size. This paper introduces Physics-Informed Extreme Learning Machines (PIELMs) as fast alternative to PINNs for solving both forward and inverse problems in financial PDEs. PIELMs replace iterative optimization with a single least-squares solve, enabling deterministic and efficient training. We benchmark PIELM on the Black-Scholes and Heston-Hull-White models for forward pricing and demonstrate its capability in inverse model calibration to recover volatility and interest rate parameters from noisy data. From experiments we observe that PIELM achieve accuracy comparable to PINNs while being up to $30\times$ faster, highlighting their potential for real-time financial modeling.
PICS in Pics: Physics Informed Contour Selection for Rapid Image Segmentation
Nov 13, 2023Abstract:Effective training of deep image segmentation models is challenging due to the need for abundant, high-quality annotations. Generating annotations is laborious and time-consuming for human experts, especially in medical image segmentation. To facilitate image annotation, we introduce Physics Informed Contour Selection (PICS) - an interpretable, physics-informed algorithm for rapid image segmentation without relying on labeled data. PICS draws inspiration from physics-informed neural networks (PINNs) and an active contour model called snake. It is fast and computationally lightweight because it employs cubic splines instead of a deep neural network as a basis function. Its training parameters are physically interpretable because they directly represent control knots of the segmentation curve. Traditional snakes involve minimization of the edge-based loss functionals by deriving the Euler-Lagrange equation followed by its numerical solution. However, PICS directly minimizes the loss functional, bypassing the Euler Lagrange equations. It is the first snake variant to minimize a region-based loss function instead of traditional edge-based loss functions. PICS uniquely models the three-dimensional (3D) segmentation process with an unsteady partial differential equation (PDE), which allows accelerated segmentation via transfer learning. To demonstrate its effectiveness, we apply PICS for 3D segmentation of the left ventricle on a publicly available cardiac dataset. While doing so, we also introduce a new convexity-preserving loss term that encodes the shape information of the left ventricle to enhance PICS's segmentation quality. Overall, PICS presents several novelties in network architecture, transfer learning, and physics-inspired losses for image segmentation, thereby showing promising outcomes and potential for further refinement.
Numerical Approximation in CFD Problems Using Physics Informed Machine Learning
Nov 01, 2021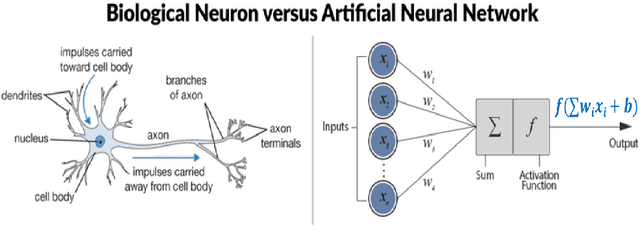
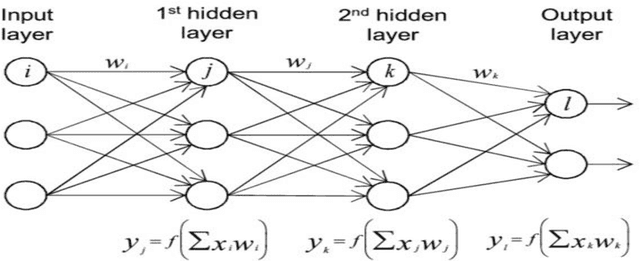
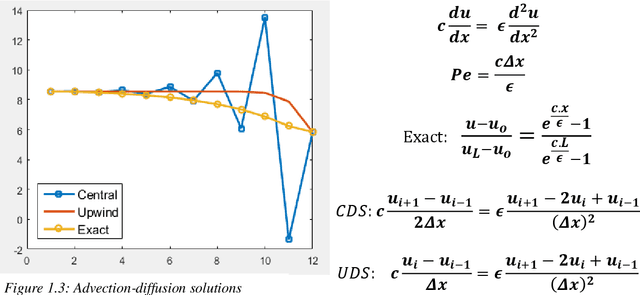
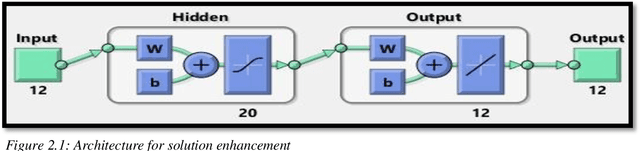
Abstract:The thesis focuses on various techniques to find an alternate approximation method that could be universally used for a wide range of CFD problems but with low computational cost and low runtime. Various techniques have been explored within the field of machine learning to gauge the utility in fulfilling the core ambition. Steady advection diffusion problem has been used as the test case to understand the level of complexity up to which a method can provide solution. Ultimately, the focus stays over physics informed machine learning techniques where solving differential equations is possible without any training with computed data. The prevalent methods by I.E. Lagaris et.al. and M. Raissi et.al are explored thoroughly. The prevalent methods cannot solve advection dominant problems. A physics informed method, called as Distributed Physics Informed Neural Network (DPINN), is proposed to solve advection dominant problems. It increases the lexibility and capability of older methods by splitting the domain and introducing other physics-based constraints as mean squared loss terms. Various experiments are done to explore the end to end possibilities with the method. Parametric study is also done to understand the behavior of the method to different tunable parameters. The method is tested over steady advection-diffusion problems and unsteady square pulse problems. Very accurate results are recorded. Extreme learning machine (ELM) is a very fast neural network algorithm at the cost of tunable parameters. The ELM based variant of the proposed model is tested over the advection-diffusion problem. ELM makes the complex optimization simpler and Since the method is non-iterative, the solution is recorded in a single shot. The ELM based variant seems to work better than the simple DPINN method. Simultaneously scope for various development in future are hinted throughout the thesis.
Abstracting Deep Neural Networks into Concept Graphs for Concept Level Interpretability
Aug 14, 2020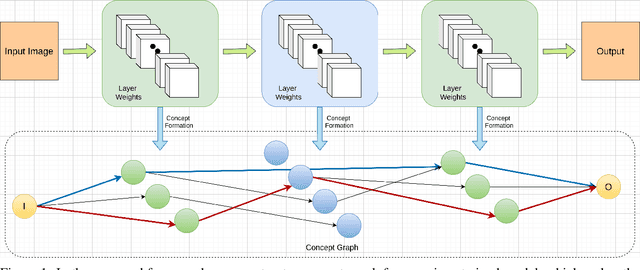

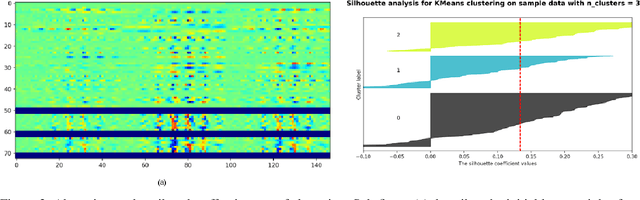
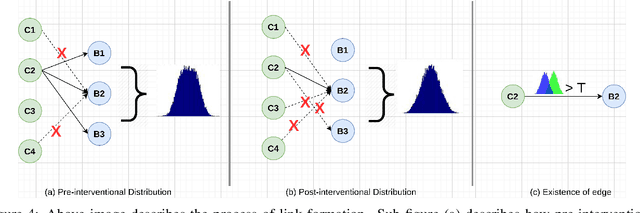
Abstract:The black-box nature of deep learning models prevents them from being completely trusted in domains like biomedicine. Most explainability techniques do not capture the concept-based reasoning that human beings follow. In this work, we attempt to understand the behavior of trained models that perform image processing tasks in the medical domain by building a graphical representation of the concepts they learn. Extracting such a graphical representation of the model's behavior on an abstract, higher conceptual level would unravel the learnings of these models and would help us to evaluate the steps taken by the model for predictions. We show the application of our proposed implementation on two biomedical problems - brain tumor segmentation and fundus image classification. We provide an alternative graphical representation of the model by formulating a \textit{concept level graph} as discussed above, which makes the problem of intervention to find active inference trails more tractable. Understanding these trails would provide an understanding of the hierarchy of the decision-making process followed by the model. [As well as overall nature of model]. Our framework is available at \url{https://github.com/koriavinash1/BioExp}
A Generalized Deep Learning Framework for Whole-Slide Image Segmentation and Analysis
Jan 01, 2020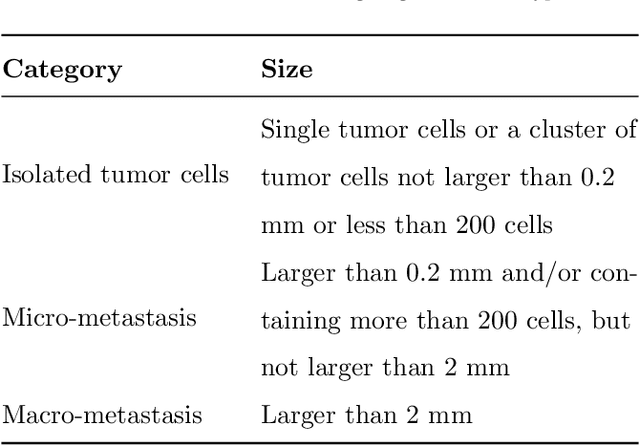
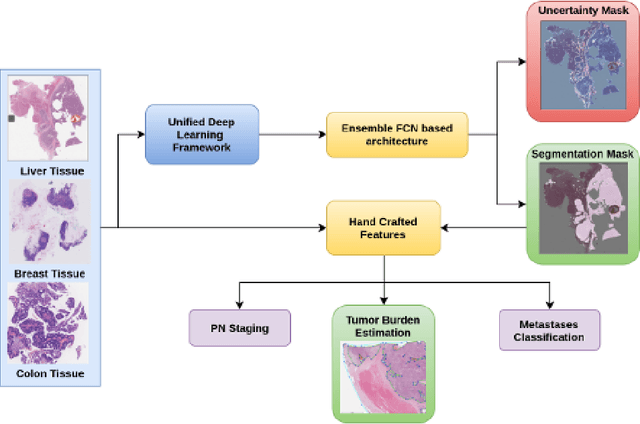
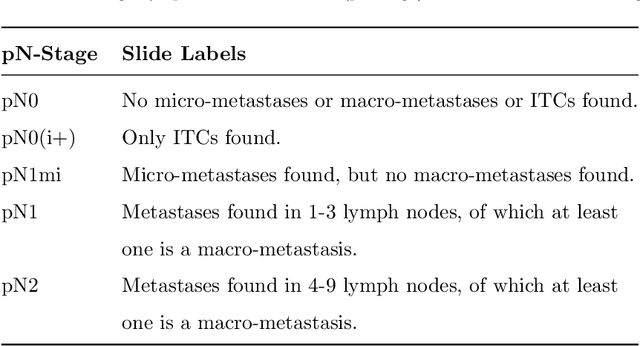
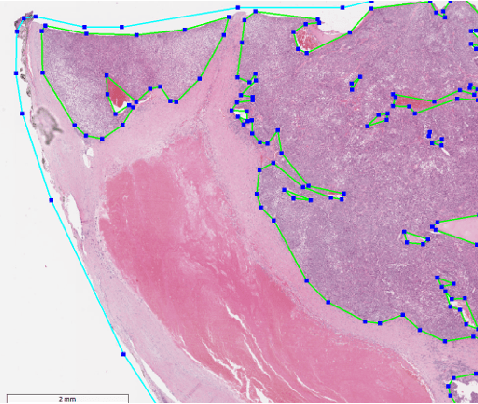
Abstract:Histopathology tissue analysis is considered the gold standard in cancer diagnosis and prognosis. Given the large size of these images and the increase in the number of potential cancer cases, an automated solution as an aid to histopathologists is highly desirable. In the recent past, deep learning-based techniques have provided state of the art results in a wide variety of image analysis tasks, including analysis of digitized slides. However, the size of images and variability in histopathology tasks makes it a challenge to develop an integrated framework for histopathology image analysis. We propose a deep learning-based framework for histopathology tissue analysis. We demonstrate the generalizability of our framework, including training and inference, on several open-source datasets, which include CAMELYON (breast cancer metastases), DigestPath (colon cancer), and PAIP (liver cancer) datasets. We discuss multiple types of uncertainties pertaining to data and model, namely aleatoric and epistemic, respectively. Simultaneously, we demonstrate our model generalization across different data distribution by evaluating some samples on TCGA data. On CAMELYON16 test data (n=139) for the task of lesion detection, the FROC score achieved was 0.86 and in the CAMELYON17 test-data (n=500) for the task of pN-staging the Cohen's kappa score achieved was 0.9090 (third in the open leaderboard). On DigestPath test data (n=212) for the task of tumor segmentation, a Dice score of 0.782 was achieved (fourth in the challenge). On PAIP test data (n=40) for the task of viable tumor segmentation, a Jaccard Index of 0.75 (third in the challenge) was achieved, and for viable tumor burden, a score of 0.633 was achieved (second in the challenge). Our entire framework and related documentation are freely available at GitHub and PyPi.
Modelling pressure-Hessian from local velocity gradients information in an incompressible turbulent flow field using deep neural networks
Nov 19, 2019
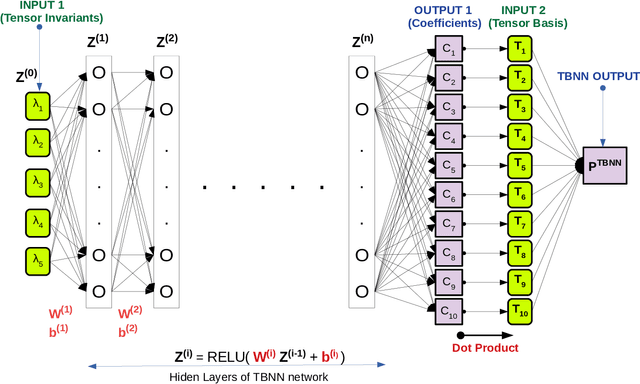
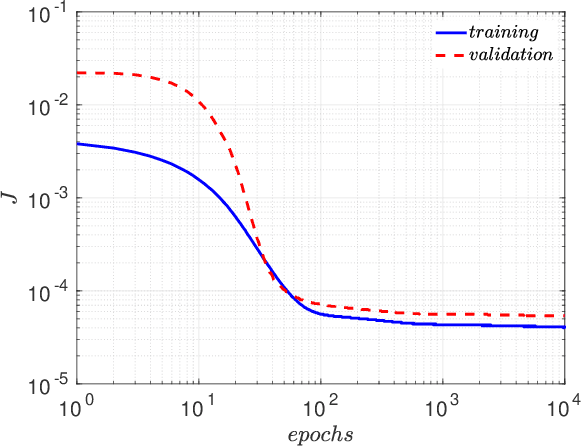

Abstract:The understanding of the dynamics of the velocity gradients in turbulent flows is critical to understanding various non-linear turbulent processes. The pressure-Hessian and the viscous-Laplacian govern the evolution of the velocity-gradients and are known to be non-local in nature. Over the years, several simplified dynamical models have been proposed that models the viscous-Laplacian and the pressure-Hessian primarily in terms of local velocity gradients information. These models can also serve as closure models for the Lagrangian PDF methods. The recent fluid deformation closure model (RFDM) has been shown to retrieve excellent one-time statistics of the viscous process. However, the pressure-Hessian modelled by the RFDM has various physical limitations. In this work, we first demonstrate the limitations of the RFDM in estimating the pressure-Hessian. Further, we employ a tensor basis neural network (TBNN) to model the pressure-Hessian from the velocity gradient tensor itself. The neural network is trained on high-resolution data obtained from direct numerical simulation (DNS) of isotropic turbulence at Reynolds number of 433 (JHU turbulence database, JHTD). The predictions made by the TBNN are tested against two different isotropic turbulence datasets at Reynolds number of 433 (JHTD) and 315 (UP Madrid turbulence database, UPMTD) and channel flow dataset at Reynolds number of 1000 (UT Texas and JHTD). The evaluation of the neural network output is made in terms of the alignment statistics of the predicted pressure-Hessian eigenvectors with the strain-rate eigenvectors for turbulent isotropic flow as well as channel flow. Our analysis of the predicted solution leads to the discovery of ten unique coefficients of the tensor basis of strain-rate and rotation-rate tensors, the linear combination over which accurately captures key alignment statistics of the pressure-Hessian tensor.
Distributed physics informed neural network for data-efficient solution to partial differential equations
Jul 21, 2019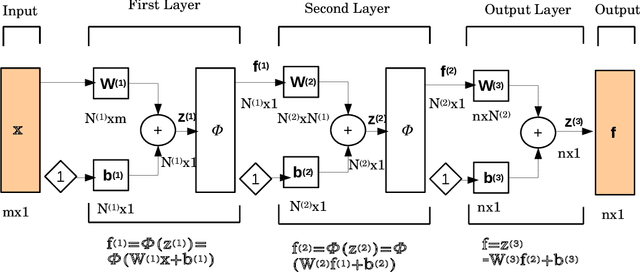



Abstract:The physics informed neural network (PINN) is evolving as a viable method to solve partial differential equations. In the recent past PINNs have been successfully tested and validated to find solutions to both linear and non-linear partial differential equations (PDEs). However, the literature lacks detailed investigation of PINNs in terms of their representation capability. In this work, we first test the original PINN method in terms of its capability to represent a complicated function. Further, to address the shortcomings of the PINN architecture, we propose a novel distributed PINN, named DPINN. We first perform a direct comparison of the proposed DPINN approach against PINN to solve a non-linear PDE (Burgers' equation). We show that DPINN not only yields a more accurate solution to the Burgers' equation, but it is found to be more data-efficient as well. At last, we employ our novel DPINN to two-dimensional steady-state Navier-Stokes equation, which is a system of non-linear PDEs. To the best of the authors' knowledge, this is the first such attempt to directly solve the Navier-Stokes equation using a physics informed neural network.
Physics Informed Extreme Learning Machine (PIELM) -- A rapid method for the numerical solution of partial differential equations
Jul 08, 2019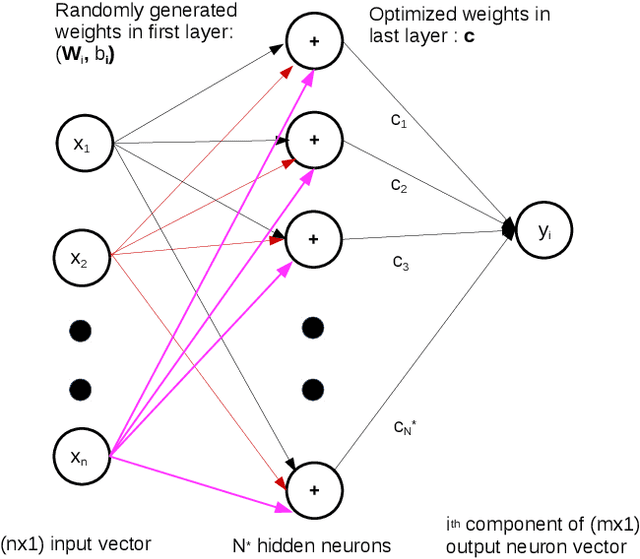
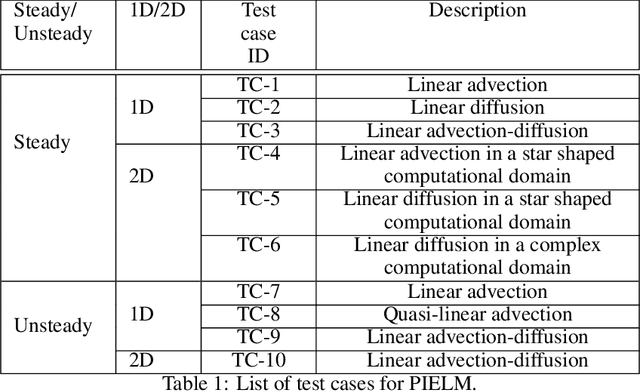
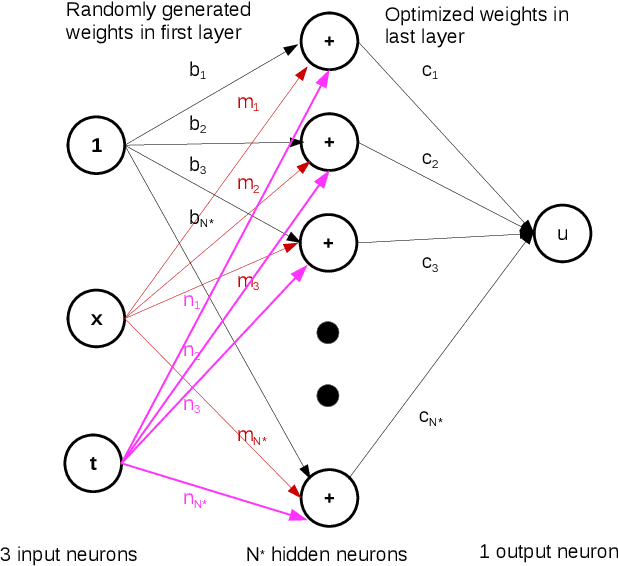

Abstract:There has been rapid progress recently on the application of deep networks to the solution of partial differential equations, collectively labelled as Physics Informed Neural Networks (PINNs). In this paper, we develop Physics Informed Extreme Learning Machine (PIELM), a rapid version of PINNs which can be applied to stationary and time dependent linear partial differential equations. We demonstrate that PIELM matches or exceeds the accuracy of PINNs on a range of problems. We also discuss the limitations of neural network based approaches, including our PIELM, in the solution of PDEs on large domains and suggest an extension, a distributed version of our algorithm -{}- DPIELM. We show that DPIELM produces excellent results comparable to conventional numerical techniques in the solution of time-dependent problems. Collectively, this work contributes towards making the use of neural networks in the solution of partial differential equations in complex domains as a competitive alternative to conventional discretization techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge