Felipe Llinares-López
Implicit Diffusion: Efficient Optimization through Stochastic Sampling
Feb 08, 2024



Abstract:We present a new algorithm to optimize distributions defined implicitly by parameterized stochastic diffusions. Doing so allows us to modify the outcome distribution of sampling processes by optimizing over their parameters. We introduce a general framework for first-order optimization of these processes, that performs jointly, in a single loop, optimization and sampling steps. This approach is inspired by recent advances in bilevel optimization and automatic implicit differentiation, leveraging the point of view of sampling as optimization over the space of probability distributions. We provide theoretical guarantees on the performance of our method, as well as experimental results demonstrating its effectiveness in real-world settings.
Learning Energy Networks with Generalized Fenchel-Young Losses
May 19, 2022



Abstract:Energy-based models, a.k.a. energy networks, perform inference by optimizing an energy function, typically parametrized by a neural network. This allows one to capture potentially complex relationships between inputs and outputs. To learn the parameters of the energy function, the solution to that optimization problem is typically fed into a loss function. The key challenge for training energy networks lies in computing loss gradients, as this typically requires argmin/argmax differentiation. In this paper, building upon a generalized notion of conjugate function, which replaces the usual bilinear pairing with a general energy function, we propose generalized Fenchel-Young losses, a natural loss construction for learning energy networks. Our losses enjoy many desirable properties and their gradients can be computed efficiently without argmin/argmax differentiation. We also prove the calibration of their excess risk in the case of linear-concave energies. We demonstrate our losses on multilabel classification and imitation learning tasks.
Efficient and Modular Implicit Differentiation
May 31, 2021
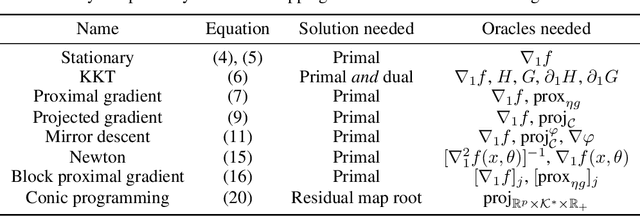
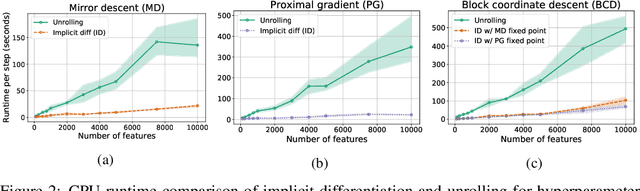

Abstract:Automatic differentiation (autodiff) has revolutionized machine learning. It allows expressing complex computations by composing elementary ones in creative ways and removes the burden of computing their derivatives by hand. More recently, differentiation of optimization problem solutions has attracted widespread attention with applications such as optimization as a layer, and in bi-level problems such as hyper-parameter optimization and meta-learning. However, the formulas for these derivatives often involve case-by-case tedious mathematical derivations. In this paper, we propose a unified, efficient and modular approach for implicit differentiation of optimization problems. In our approach, the user defines (in Python in the case of our implementation) a function $F$ capturing the optimality conditions of the problem to be differentiated. Once this is done, we leverage autodiff of $F$ and implicit differentiation to automatically differentiate the optimization problem. Our approach thus combines the benefits of implicit differentiation and autodiff. It is efficient as it can be added on top of any state-of-the-art solver and modular as the optimality condition specification is decoupled from the implicit differentiation mechanism. We show that seemingly simple principles allow to recover many recently proposed implicit differentiation methods and create new ones easily. We demonstrate the ease of formulating and solving bi-level optimization problems using our framework. We also showcase an application to the sensitivity analysis of molecular dynamics.
Graph Kernels: State-of-the-Art and Future Challenges
Nov 10, 2020



Abstract:Graph-structured data are an integral part of many application domains, including chemoinformatics, computational biology, neuroimaging, and social network analysis. Over the last two decades, numerous graph kernels, i.e. kernel functions between graphs, have been proposed to solve the problem of assessing the similarity between graphs, thereby making it possible to perform predictions in both classification and regression settings. This manuscript provides a review of existing graph kernels, their applications, software plus data resources, and an empirical comparison of state-of-the-art graph kernels.
Wasserstein Weisfeiler-Lehman Graph Kernels
Jun 04, 2019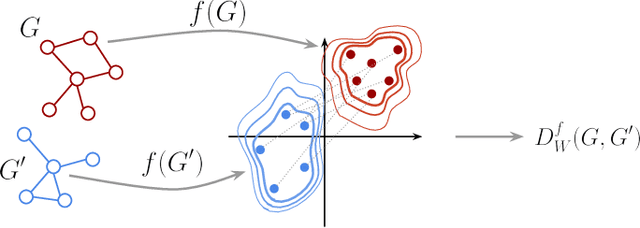
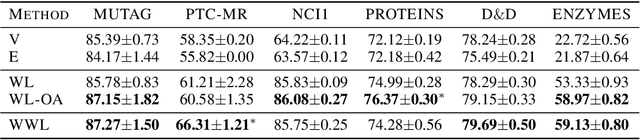
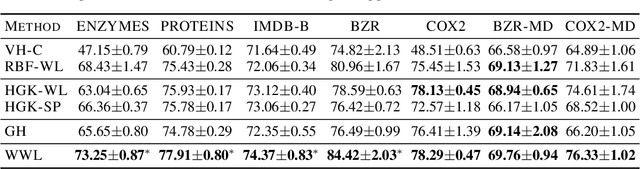
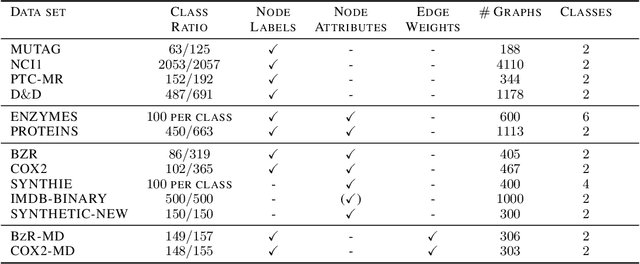
Abstract:Graph kernels are an instance of the class of $\mathcal{R}$-Convolution kernels, which measure the similarity of objects by comparing their substructures. Despite their empirical success, most graph kernels use a naive aggregation of the final set of substructures, usually a sum or average, thereby potentially discarding valuable information about the distribution of individual components. Furthermore, only a limited instance of these approaches can be extended to continuously attributed graphs. We propose a novel method that relies on the Wasserstein distance between the node feature vector distributions of two graphs, which allows to find subtler differences in data sets by considering graphs as high-dimensional objects, rather than simple means. We further propose a Weisfeiler-Lehman inspired embedding scheme for graphs with continuous node attributes and weighted edges, enhance it with the computed Wasserstein distance, and thus improve the state-of-the-art prediction performance on several graph classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge