Elisabetta Ghisu
Graph Kernels: State-of-the-Art and Future Challenges
Nov 10, 2020



Abstract:Graph-structured data are an integral part of many application domains, including chemoinformatics, computational biology, neuroimaging, and social network analysis. Over the last two decades, numerous graph kernels, i.e. kernel functions between graphs, have been proposed to solve the problem of assessing the similarity between graphs, thereby making it possible to perform predictions in both classification and regression settings. This manuscript provides a review of existing graph kernels, their applications, software plus data resources, and an empirical comparison of state-of-the-art graph kernels.
Wasserstein Weisfeiler-Lehman Graph Kernels
Jun 04, 2019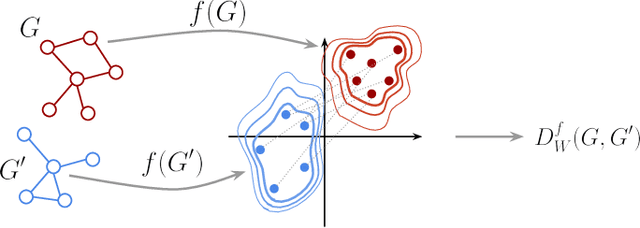
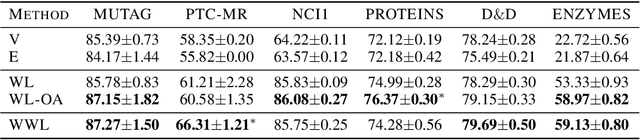
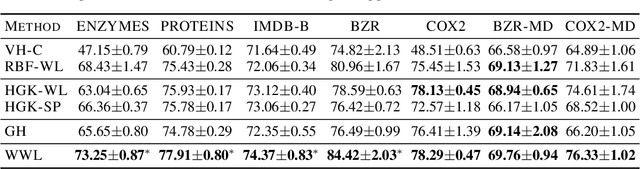
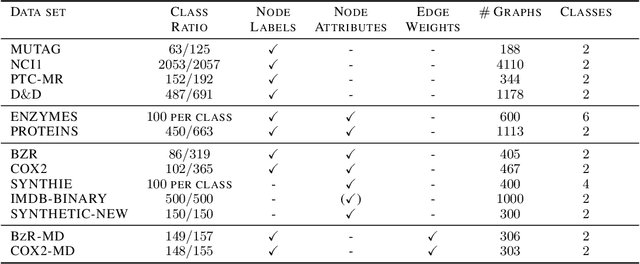
Abstract:Graph kernels are an instance of the class of $\mathcal{R}$-Convolution kernels, which measure the similarity of objects by comparing their substructures. Despite their empirical success, most graph kernels use a naive aggregation of the final set of substructures, usually a sum or average, thereby potentially discarding valuable information about the distribution of individual components. Furthermore, only a limited instance of these approaches can be extended to continuously attributed graphs. We propose a novel method that relies on the Wasserstein distance between the node feature vector distributions of two graphs, which allows to find subtler differences in data sets by considering graphs as high-dimensional objects, rather than simple means. We further propose a Weisfeiler-Lehman inspired embedding scheme for graphs with continuous node attributes and weighted edges, enhance it with the computed Wasserstein distance, and thus improve the state-of-the-art prediction performance on several graph classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge