Fabio Hellmann
The STOIC2021 COVID-19 AI challenge: applying reusable training methodologies to private data
Jun 25, 2023Abstract:Challenges drive the state-of-the-art of automated medical image analysis. The quantity of public training data that they provide can limit the performance of their solutions. Public access to the training methodology for these solutions remains absent. This study implements the Type Three (T3) challenge format, which allows for training solutions on private data and guarantees reusable training methodologies. With T3, challenge organizers train a codebase provided by the participants on sequestered training data. T3 was implemented in the STOIC2021 challenge, with the goal of predicting from a computed tomography (CT) scan whether subjects had a severe COVID-19 infection, defined as intubation or death within one month. STOIC2021 consisted of a Qualification phase, where participants developed challenge solutions using 2000 publicly available CT scans, and a Final phase, where participants submitted their training methodologies with which solutions were trained on CT scans of 9724 subjects. The organizers successfully trained six of the eight Final phase submissions. The submitted codebases for training and running inference were released publicly. The winning solution obtained an area under the receiver operating characteristic curve for discerning between severe and non-severe COVID-19 of 0.815. The Final phase solutions of all finalists improved upon their Qualification phase solutions.HSUXJM-TNZF9CHSUXJM-TNZF9C
Towards Automated COVID-19 Presence and Severity Classification
May 15, 2023Abstract:COVID-19 presence classification and severity prediction via (3D) thorax computed tomography scans have become important tasks in recent times. Especially for capacity planning of intensive care units, predicting the future severity of a COVID-19 patient is crucial. The presented approach follows state-of-theart techniques to aid medical professionals in these situations. It comprises an ensemble learning strategy via 5-fold cross-validation that includes transfer learning and combines pre-trained 3D-versions of ResNet34 and DenseNet121 for COVID19 classification and severity prediction respectively. Further, domain-specific preprocessing was applied to optimize model performance. In addition, medical information like the infection-lung-ratio, patient age, and sex were included. The presented model achieves an AUC of 79.0% to predict COVID-19 severity, and 83.7% AUC to classify the presence of an infection, which is comparable with other currently popular methods. This approach is implemented using the AUCMEDI framework and relies on well-known network architectures to ensure robustness and reproducibility.
GANonymization: A GAN-based Face Anonymization Framework for Preserving Emotional Expressions
May 03, 2023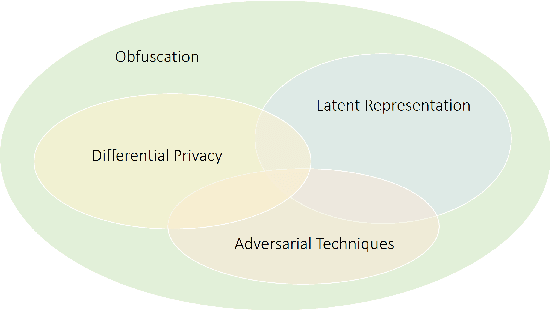

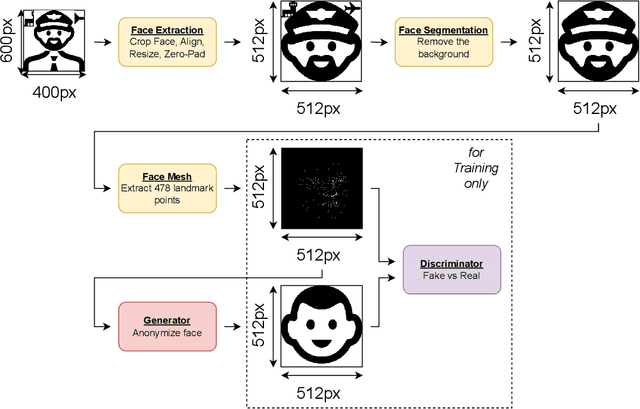

Abstract:In recent years, the increasing availability of personal data has raised concerns regarding privacy and security. One of the critical processes to address these concerns is data anonymization, which aims to protect individual privacy and prevent the release of sensitive information. This research focuses on the importance of face anonymization. Therefore, we introduce GANonymization, a novel face anonymization framework with facial expression-preserving abilities. Our approach is based on a high-level representation of a face which is synthesized into an anonymized version based on a generative adversarial network (GAN). The effectiveness of the approach was assessed by evaluating its performance in removing identifiable facial attributes to increase the anonymity of the given individual face. Additionally, the performance of preserving facial expressions was evaluated on several affect recognition datasets and outperformed the state-of-the-art method in most categories. Finally, our approach was analyzed for its ability to remove various facial traits, such as jewelry, hair color, and multiple others. Here, it demonstrated reliable performance in removing these attributes. Our results suggest that GANonymization is a promising approach for anonymizing faces while preserving facial expressions.
Improving Deep Facial Phenotyping for Ultra-rare Disorder Verification Using Model Ensembles
Nov 12, 2022



Abstract:Rare genetic disorders affect more than 6% of the global population. Reaching a diagnosis is challenging because rare disorders are very diverse. Many disorders have recognizable facial features that are hints for clinicians to diagnose patients. Previous work, such as GestaltMatcher, utilized representation vectors produced by a DCNN similar to AlexNet to match patients in high-dimensional feature space to support "unseen" ultra-rare disorders. However, the architecture and dataset used for transfer learning in GestaltMatcher have become outdated. Moreover, a way to train the model for generating better representation vectors for unseen ultra-rare disorders has not yet been studied. Because of the overall scarcity of patients with ultra-rare disorders, it is infeasible to directly train a model on them. Therefore, we first analyzed the influence of replacing GestaltMatcher DCNN with a state-of-the-art face recognition approach, iResNet with ArcFace. Additionally, we experimented with different face recognition datasets for transfer learning. Furthermore, we proposed test-time augmentation, and model ensembles that mix general face verification models and models specific for verifying disorders to improve the disorder verification accuracy of unseen ultra-rare disorders. Our proposed ensemble model achieves state-of-the-art performance on both seen and unseen disorders.
Few-Shot Meta Learning for Recognizing Facial Phenotypes of Genetic Disorders
Oct 23, 2022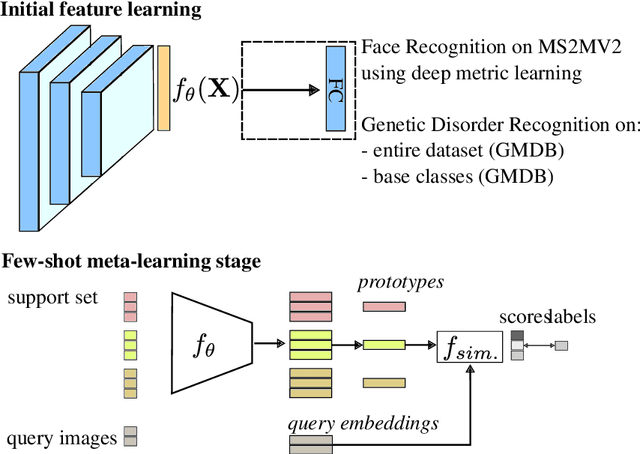
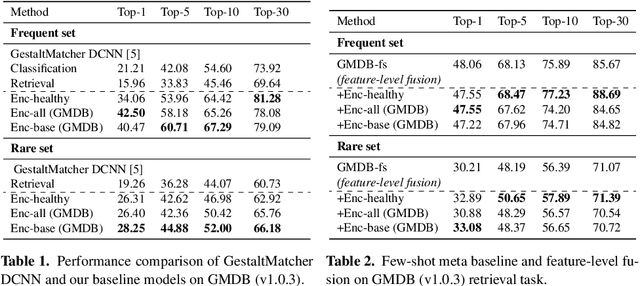
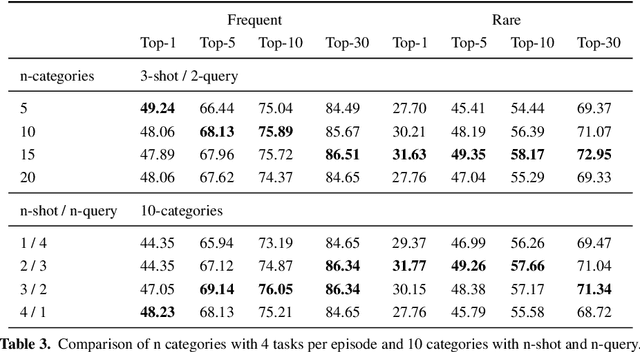
Abstract:Computer vision-based methods have valuable use cases in precision medicine, and recognizing facial phenotypes of genetic disorders is one of them. Many genetic disorders are known to affect faces' visual appearance and geometry. Automated classification and similarity retrieval aid physicians in decision-making to diagnose possible genetic conditions as early as possible. Previous work has addressed the problem as a classification problem and used deep learning methods. The challenging issue in practice is the sparse label distribution and huge class imbalances across categories. Furthermore, most disorders have few labeled samples in training sets, making representation learning and generalization essential to acquiring a reliable feature descriptor. In this study, we used a facial recognition model trained on a large corpus of healthy individuals as a pre-task and transferred it to facial phenotype recognition. Furthermore, we created simple baselines of few-shot meta-learning methods to improve our base feature descriptor. Our quantitative results on GestaltMatcher Database show that our CNN baseline surpasses previous works, including GestaltMatcher, and few-shot meta-learning strategies improve retrieval performance in frequent and rare classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge