Tzung-Chien Hsieh
Multimodal Machine Learning Combining Facial Images and Clinical Texts Improves Diagnosis of Rare Genetic Diseases
Dec 23, 2023Abstract:Individuals with suspected rare genetic disorders often undergo multiple clinical evaluations, imaging studies, laboratory tests and genetic tests, to find a possible answer over a prolonged period of multiple years. Addressing this diagnostic odyssey thus have substantial clinical, psychosocial, and economic benefits. Many rare genetic diseases have distinctive facial features, which can be used by artificial intelligence algorithms to facilitate clinical diagnosis, in prioritizing candidate diseases to be further examined by lab tests or genetic assays, or in helping the phenotype-driven reinterpretation of genome/exome sequencing data. However, existing methods using frontal facial photo were built on conventional Convolutional Neural Networks (CNNs), rely exclusively on facial images, and cannot capture non-facial phenotypic traits and demographic information essential for guiding accurate diagnoses. Here we introduce GestaltMML, a multimodal machine learning (MML) approach solely based on the Transformer architecture. It integrates the facial images, demographic information (age, sex, ethnicity), and clinical notes of patients to improve prediction accuracy. Furthermore, we also introduce GestaltGPT, a GPT-based methodology with few-short learning capacities that exclusively harnesses textual inputs using a range of large language models (LLMs) including Llama 2, GPT-J and Falcon. We evaluated these methods on a diverse range of datasets, including 449 diseases from the GestaltMatcher Database, several in-house datasets on Beckwith-Wiedemann syndrome, Sotos syndrome, NAA10-related syndrome (neurodevelopmental syndrome) and others. Our results suggest that GestaltMML/GestaltGPT effectively incorporate multiple modalities of data, greatly narrow down candidate genetic diagnosis of rare diseases, and may facilitate the reinterpretation of genome/exome sequencing data.
GANonymization: A GAN-based Face Anonymization Framework for Preserving Emotional Expressions
May 03, 2023

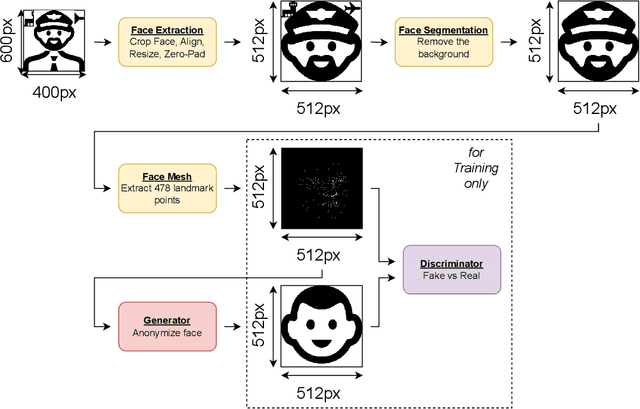

Abstract:In recent years, the increasing availability of personal data has raised concerns regarding privacy and security. One of the critical processes to address these concerns is data anonymization, which aims to protect individual privacy and prevent the release of sensitive information. This research focuses on the importance of face anonymization. Therefore, we introduce GANonymization, a novel face anonymization framework with facial expression-preserving abilities. Our approach is based on a high-level representation of a face which is synthesized into an anonymized version based on a generative adversarial network (GAN). The effectiveness of the approach was assessed by evaluating its performance in removing identifiable facial attributes to increase the anonymity of the given individual face. Additionally, the performance of preserving facial expressions was evaluated on several affect recognition datasets and outperformed the state-of-the-art method in most categories. Finally, our approach was analyzed for its ability to remove various facial traits, such as jewelry, hair color, and multiple others. Here, it demonstrated reliable performance in removing these attributes. Our results suggest that GANonymization is a promising approach for anonymizing faces while preserving facial expressions.
Improving Deep Facial Phenotyping for Ultra-rare Disorder Verification Using Model Ensembles
Nov 12, 2022



Abstract:Rare genetic disorders affect more than 6% of the global population. Reaching a diagnosis is challenging because rare disorders are very diverse. Many disorders have recognizable facial features that are hints for clinicians to diagnose patients. Previous work, such as GestaltMatcher, utilized representation vectors produced by a DCNN similar to AlexNet to match patients in high-dimensional feature space to support "unseen" ultra-rare disorders. However, the architecture and dataset used for transfer learning in GestaltMatcher have become outdated. Moreover, a way to train the model for generating better representation vectors for unseen ultra-rare disorders has not yet been studied. Because of the overall scarcity of patients with ultra-rare disorders, it is infeasible to directly train a model on them. Therefore, we first analyzed the influence of replacing GestaltMatcher DCNN with a state-of-the-art face recognition approach, iResNet with ArcFace. Additionally, we experimented with different face recognition datasets for transfer learning. Furthermore, we proposed test-time augmentation, and model ensembles that mix general face verification models and models specific for verifying disorders to improve the disorder verification accuracy of unseen ultra-rare disorders. Our proposed ensemble model achieves state-of-the-art performance on both seen and unseen disorders.
Few-Shot Meta Learning for Recognizing Facial Phenotypes of Genetic Disorders
Oct 23, 2022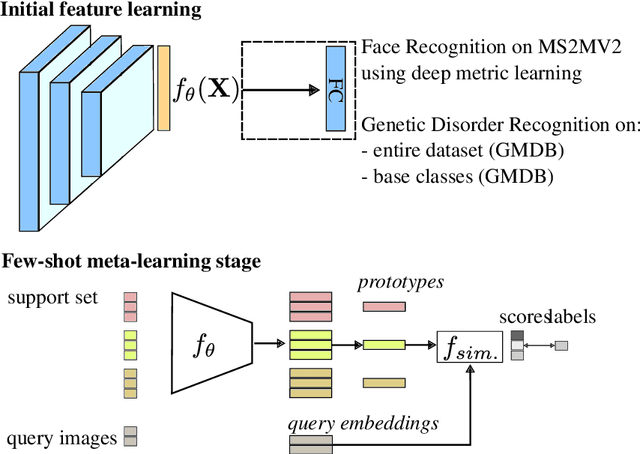
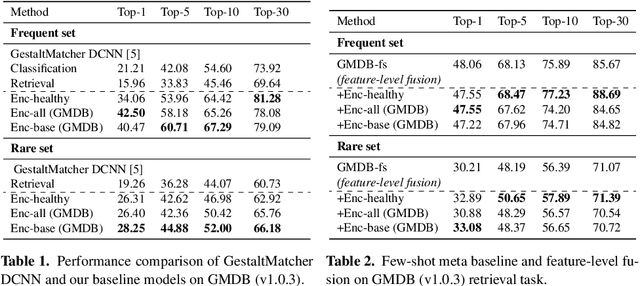
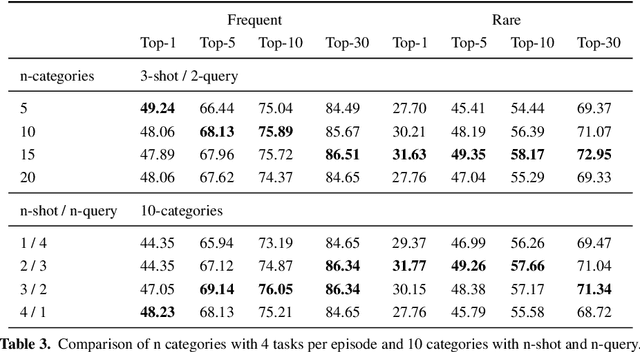
Abstract:Computer vision-based methods have valuable use cases in precision medicine, and recognizing facial phenotypes of genetic disorders is one of them. Many genetic disorders are known to affect faces' visual appearance and geometry. Automated classification and similarity retrieval aid physicians in decision-making to diagnose possible genetic conditions as early as possible. Previous work has addressed the problem as a classification problem and used deep learning methods. The challenging issue in practice is the sparse label distribution and huge class imbalances across categories. Furthermore, most disorders have few labeled samples in training sets, making representation learning and generalization essential to acquiring a reliable feature descriptor. In this study, we used a facial recognition model trained on a large corpus of healthy individuals as a pre-task and transferred it to facial phenotype recognition. Furthermore, we created simple baselines of few-shot meta-learning methods to improve our base feature descriptor. Our quantitative results on GestaltMatcher Database show that our CNN baseline surpasses previous works, including GestaltMatcher, and few-shot meta-learning strategies improve retrieval performance in frequent and rare classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge