Diego Mesquita
Boosted GFlowNets: Improving Exploration via Sequential Learning
Nov 12, 2025Abstract:Generative Flow Networks (GFlowNets) are powerful samplers for compositional objects that, by design, sample proportionally to a given non-negative reward. Nonetheless, in practice, they often struggle to explore the reward landscape evenly: trajectories toward easy-to-reach regions dominate training, while hard-to-reach modes receive vanishing or uninformative gradients, leading to poor coverage of high-reward areas. We address this imbalance with Boosted GFlowNets, a method that sequentially trains an ensemble of GFlowNets, each optimizing a residual reward that compensates for the mass already captured by previous models. This residual principle reactivates learning signals in underexplored regions and, under mild assumptions, ensures a monotone non-degradation property: adding boosters cannot worsen the learned distribution and typically improves it. Empirically, Boosted GFlowNets achieve substantially better exploration and sample diversity on multimodal synthetic benchmarks and peptide design tasks, while preserving the stability and simplicity of standard trajectory-balance training.
On the Identifiability of Tensor Ranks via Prior Predictive Matching
Oct 16, 2025Abstract:Selecting the latent dimensions (ranks) in tensor factorization is a central challenge that often relies on heuristic methods. This paper introduces a rigorous approach to determine rank identifiability in probabilistic tensor models, based on prior predictive moment matching. We transform a set of moment matching conditions into a log-linear system of equations in terms of marginal moments, prior hyperparameters, and ranks; establishing an equivalence between rank identifiability and the solvability of such system. We apply this framework to four foundational tensor-models, demonstrating that the linear structure of the PARAFAC/CP model, the chain structure of the Tensor Train model, and the closed-loop structure of the Tensor Ring model yield solvable systems, making their ranks identifiable. In contrast, we prove that the symmetric topology of the Tucker model leads to an underdetermined system, rendering the ranks unidentifiable by this method. For the identifiable models, we derive explicit closed-form rank estimators based on the moments of observed data only. We empirically validate these estimators and evaluate the robustness of the proposal.
Differentially Private Selection using Smooth Sensitivity
Apr 10, 2025Abstract:Differentially private selection mechanisms offer strong privacy guarantees for queries aiming to identify the top-scoring element r from a finite set R, based on a dataset-dependent utility function. While selection queries are fundamental in data science, few mechanisms effectively ensure their privacy. Furthermore, most approaches rely on global sensitivity to achieve differential privacy (DP), which can introduce excessive noise and impair downstream inferences. To address this limitation, we propose the Smooth Noisy Max (SNM) mechanism, which leverages smooth sensitivity to yield provably tighter (upper bounds on) expected errors compared to global sensitivity-based methods. Empirical results demonstrate that SNM is more accurate than state-of-the-art differentially private selection methods in three applications: percentile selection, greedy decision trees, and random forests.
Streaming Bayes GFlowNets
Nov 08, 2024



Abstract:Bayes' rule naturally allows for inference refinement in a streaming fashion, without the need to recompute posteriors from scratch whenever new data arrives. In principle, Bayesian streaming is straightforward: we update our prior with the available data and use the resulting posterior as a prior when processing the next data chunk. In practice, however, this recipe entails i) approximating an intractable posterior at each time step; and ii) encapsulating results appropriately to allow for posterior propagation. For continuous state spaces, variational inference (VI) is particularly convenient due to its scalability and the tractability of variational posteriors. For discrete state spaces, however, state-of-the-art VI results in analytically intractable approximations that are ill-suited for streaming settings. To enable streaming Bayesian inference over discrete parameter spaces, we propose streaming Bayes GFlowNets (abbreviated as SB-GFlowNets) by leveraging the recently proposed GFlowNets -- a powerful class of amortized samplers for discrete compositional objects. Notably, SB-GFlowNet approximates the initial posterior using a standard GFlowNet and subsequently updates it using a tailored procedure that requires only the newly observed data. Our case studies in linear preference learning and phylogenetic inference showcase the effectiveness of SB-GFlowNets in sampling from an unnormalized posterior in a streaming setting. As expected, we also observe that SB-GFlowNets is significantly faster than repeatedly training a GFlowNet from scratch to sample from the full posterior.
On Divergence Measures for Training GFlowNets
Oct 12, 2024Abstract:Generative Flow Networks (GFlowNets) are amortized inference models designed to sample from unnormalized distributions over composable objects, with applications in generative modeling for tasks in fields such as causal discovery, NLP, and drug discovery. Traditionally, the training procedure for GFlowNets seeks to minimize the expected log-squared difference between a proposal (forward policy) and a target (backward policy) distribution, which enforces certain flow-matching conditions. While this training procedure is closely related to variational inference (VI), directly attempting standard Kullback-Leibler (KL) divergence minimization can lead to proven biased and potentially high-variance estimators. Therefore, we first review four divergence measures, namely, Renyi-$\alpha$'s, Tsallis-$\alpha$'s, reverse and forward KL's, and design statistically efficient estimators for their stochastic gradients in the context of training GFlowNets. Then, we verify that properly minimizing these divergences yields a provably correct and empirically effective training scheme, often leading to significantly faster convergence than previously proposed optimization. To achieve this, we design control variates based on the REINFORCE leave-one-out and score-matching estimators to reduce the variance of the learning objectives' gradients. Our work contributes by narrowing the gap between GFlowNets training and generalized variational approximations, paving the way for algorithmic ideas informed by the divergence minimization viewpoint.
Embarrassingly Parallel GFlowNets
Jun 05, 2024



Abstract:GFlowNets are a promising alternative to MCMC sampling for discrete compositional random variables. Training GFlowNets requires repeated evaluations of the unnormalized target distribution or reward function. However, for large-scale posterior sampling, this may be prohibitive since it incurs traversing the data several times. Moreover, if the data are distributed across clients, employing standard GFlowNets leads to intensive client-server communication. To alleviate both these issues, we propose embarrassingly parallel GFlowNet (EP-GFlowNet). EP-GFlowNet is a provably correct divide-and-conquer method to sample from product distributions of the form $R(\cdot) \propto R_1(\cdot) ... R_N(\cdot)$ -- e.g., in parallel or federated Bayes, where each $R_n$ is a local posterior defined on a data partition. First, in parallel, we train a local GFlowNet targeting each $R_n$ and send the resulting models to the server. Then, the server learns a global GFlowNet by enforcing our newly proposed \emph{aggregating balance} condition, requiring a single communication step. Importantly, EP-GFlowNets can also be applied to multi-objective optimization and model reuse. Our experiments illustrate the EP-GFlowNets's effectiveness on many tasks, including parallel Bayesian phylogenetics, multi-objective multiset, sequence generation, and federated Bayesian structure learning.
In-n-Out: Calibrating Graph Neural Networks for Link Prediction
Mar 08, 2024Abstract:Deep neural networks are notoriously miscalibrated, i.e., their outputs do not reflect the true probability of the event we aim to predict. While networks for tabular or image data are usually overconfident, recent works have shown that graph neural networks (GNNs) show the opposite behavior for node-level classification. But what happens when we are predicting links? We show that, in this case, GNNs often exhibit a mixed behavior. More specifically, they may be overconfident in negative predictions while being underconfident in positive ones. Based on this observation, we propose IN-N-OUT, the first-ever method to calibrate GNNs for link prediction. IN-N-OUT is based on two simple intuitions: i) attributing true/false labels to an edge while respecting a GNNs prediction should cause but small fluctuations in that edge's embedding; and, conversely, ii) if we label that same edge contradicting our GNN, embeddings should change more substantially. An extensive experimental campaign shows that IN-N-OUT significantly improves the calibration of GNNs in link prediction, consistently outperforming the baselines available -- which are not designed for this specific task.
Thin and Deep Gaussian Processes
Oct 17, 2023Abstract:Gaussian processes (GPs) can provide a principled approach to uncertainty quantification with easy-to-interpret kernel hyperparameters, such as the lengthscale, which controls the correlation distance of function values. However, selecting an appropriate kernel can be challenging. Deep GPs avoid manual kernel engineering by successively parameterizing kernels with GP layers, allowing them to learn low-dimensional embeddings of the inputs that explain the output data. Following the architecture of deep neural networks, the most common deep GPs warp the input space layer-by-layer but lose all the interpretability of shallow GPs. An alternative construction is to successively parameterize the lengthscale of a kernel, improving the interpretability but ultimately giving away the notion of learning lower-dimensional embeddings. Unfortunately, both methods are susceptible to particular pathologies which may hinder fitting and limit their interpretability. This work proposes a novel synthesis of both previous approaches: Thin and Deep GP (TDGP). Each TDGP layer defines locally linear transformations of the original input data maintaining the concept of latent embeddings while also retaining the interpretation of lengthscales of a kernel. Moreover, unlike the prior solutions, TDGP induces non-pathological manifolds that admit learning lower-dimensional representations. We show with theoretical and experimental results that i) TDGP is, unlike previous models, tailored to specifically discover lower-dimensional manifolds in the input data, ii) TDGP behaves well when increasing the number of layers, and iii) TDGP performs well in standard benchmark datasets.
Human-in-the-Loop Causal Discovery under Latent Confounding using Ancestral GFlowNets
Sep 21, 2023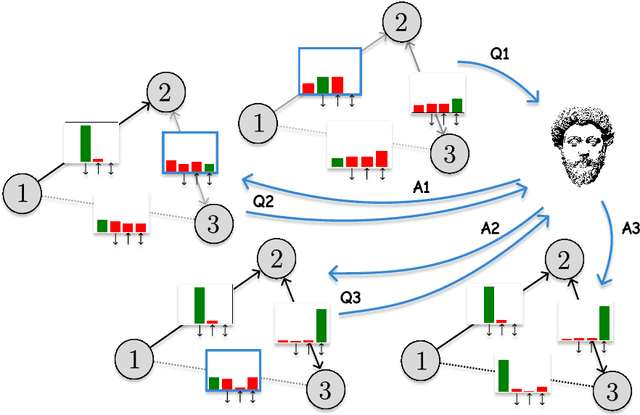
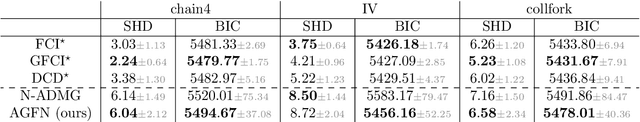
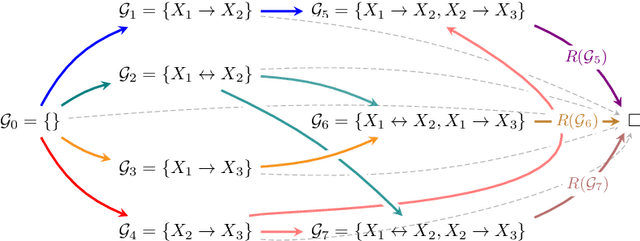
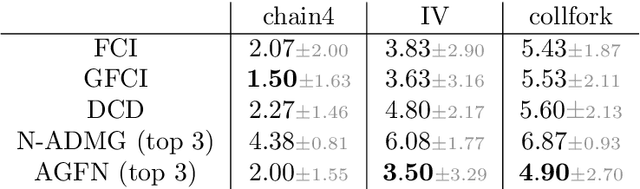
Abstract:Structure learning is the crux of causal inference. Notably, causal discovery (CD) algorithms are brittle when data is scarce, possibly inferring imprecise causal relations that contradict expert knowledge -- especially when considering latent confounders. To aggravate the issue, most CD methods do not provide uncertainty estimates, making it hard for users to interpret results and improve the inference process. Surprisingly, while CD is a human-centered affair, no works have focused on building methods that both 1) output uncertainty estimates that can be verified by experts and 2) interact with those experts to iteratively refine CD. To solve these issues, we start by proposing to sample (causal) ancestral graphs proportionally to a belief distribution based on a score function, such as the Bayesian information criterion (BIC), using generative flow networks. Then, we leverage the diversity in candidate graphs and introduce an optimal experimental design to iteratively probe the expert about the relations among variables, effectively reducing the uncertainty of our belief over ancestral graphs. Finally, we update our samples to incorporate human feedback via importance sampling. Importantly, our method does not require causal sufficiency (i.e., unobserved confounders may exist). Experiments with synthetic observational data show that our method can accurately sample from distributions over ancestral graphs and that we can greatly improve inference quality with human aid.
Locking and Quacking: Stacking Bayesian model predictions by log-pooling and superposition
May 12, 2023



Abstract:Combining predictions from different models is a central problem in Bayesian inference and machine learning more broadly. Currently, these predictive distributions are almost exclusively combined using linear mixtures such as Bayesian model averaging, Bayesian stacking, and mixture of experts. Such linear mixtures impose idiosyncrasies that might be undesirable for some applications, such as multi-modality. While there exist alternative strategies (e.g. geometric bridge or superposition), optimising their parameters usually involves computing an intractable normalising constant repeatedly. We present two novel Bayesian model combination tools. These are generalisations of model stacking, but combine posterior densities by log-linear pooling (locking) and quantum superposition (quacking). To optimise model weights while avoiding the burden of normalising constants, we investigate the Hyvarinen score of the combined posterior predictions. We demonstrate locking with an illustrative example and discuss its practical application with importance sampling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge