Dominik Heider
MetaMP: Seamless Metadata Enrichment and AI Application Framework for Enhanced Membrane Protein Visualization and Analysis
Oct 06, 2025Abstract:Structural biology has made significant progress in determining membrane proteins, leading to a remarkable increase in the number of available structures in dedicated databases. The inherent complexity of membrane protein structures, coupled with challenges such as missing data, inconsistencies, and computational barriers from disparate sources, underscores the need for improved database integration. To address this gap, we present MetaMP, a framework that unifies membrane-protein databases within a web application and uses machine learning for classification. MetaMP improves data quality by enriching metadata, offering a user-friendly interface, and providing eight interactive views for streamlined exploration. MetaMP was effective across tasks of varying difficulty, demonstrating advantages across different levels without compromising speed or accuracy, according to user evaluations. Moreover, MetaMP supports essential functions such as structure classification and outlier detection. We present three practical applications of Artificial Intelligence (AI) in membrane protein research: predicting transmembrane segments, reconciling legacy databases, and classifying structures with explainable AI support. In a validation focused on statistics, MetaMP resolved 77% of data discrepancies and accurately predicted the class of newly identified membrane proteins 98% of the time and overtook expert curation. Altogether, MetaMP is a much-needed resource that harmonizes current knowledge and empowers AI-driven exploration of membrane-protein architecture.
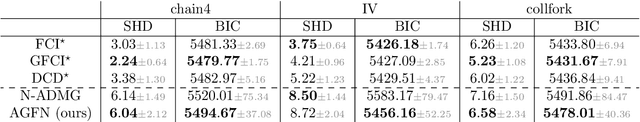
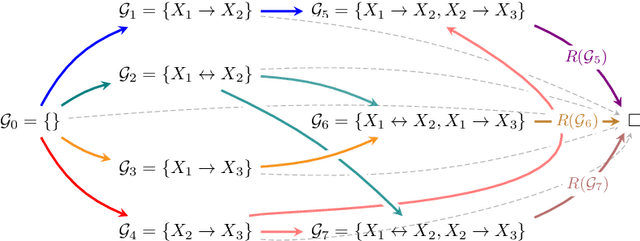
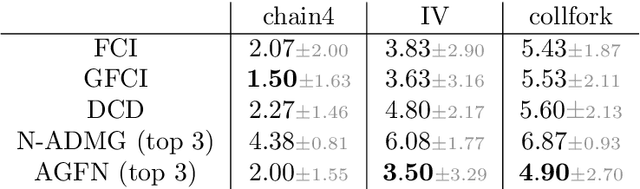
dcFCI: Robust Causal Discovery Under Latent Confounding, Unfaithfulness, and Mixed Data
May 10, 2025Abstract:Causal discovery is central to inferring causal relationships from observational data. In the presence of latent confounding, algorithms such as Fast Causal Inference (FCI) learn a Partial Ancestral Graph (PAG) representing the true model's Markov Equivalence Class. However, their correctness critically depends on empirical faithfulness, the assumption that observed (in)dependencies perfectly reflect those of the underlying causal model, which often fails in practice due to limited sample sizes. To address this, we introduce the first nonparametric score to assess a PAG's compatibility with observed data, even with mixed variable types. This score is both necessary and sufficient to characterize structural uncertainty and distinguish between distinct PAGs. We then propose data-compatible FCI (dcFCI), the first hybrid causal discovery algorithm to jointly address latent confounding, empirical unfaithfulness, and mixed data types. dcFCI integrates our score into an (Anytime)FCI-guided search that systematically explores, ranks, and validates candidate PAGs. Experiments on synthetic and real-world scenarios demonstrate that dcFCI significantly outperforms state-of-the-art methods, often recovering the true PAG even in small and heterogeneous datasets. Examining top-ranked PAGs further provides valuable insights into structural uncertainty, supporting more robust and informed causal reasoning and decision-making.
Human-in-the-Loop Causal Discovery under Latent Confounding using Ancestral GFlowNets
Sep 21, 2023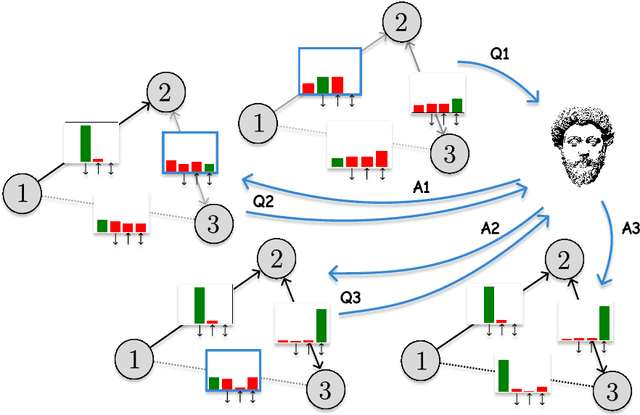
Abstract:Structure learning is the crux of causal inference. Notably, causal discovery (CD) algorithms are brittle when data is scarce, possibly inferring imprecise causal relations that contradict expert knowledge -- especially when considering latent confounders. To aggravate the issue, most CD methods do not provide uncertainty estimates, making it hard for users to interpret results and improve the inference process. Surprisingly, while CD is a human-centered affair, no works have focused on building methods that both 1) output uncertainty estimates that can be verified by experts and 2) interact with those experts to iteratively refine CD. To solve these issues, we start by proposing to sample (causal) ancestral graphs proportionally to a belief distribution based on a score function, such as the Bayesian information criterion (BIC), using generative flow networks. Then, we leverage the diversity in candidate graphs and introduce an optimal experimental design to iteratively probe the expert about the relations among variables, effectively reducing the uncertainty of our belief over ancestral graphs. Finally, we update our samples to incorporate human feedback via importance sampling. Importantly, our method does not require causal sufficiency (i.e., unobserved confounders may exist). Experiments with synthetic observational data show that our method can accurately sample from distributions over ancestral graphs and that we can greatly improve inference quality with human aid.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
May 12, 2021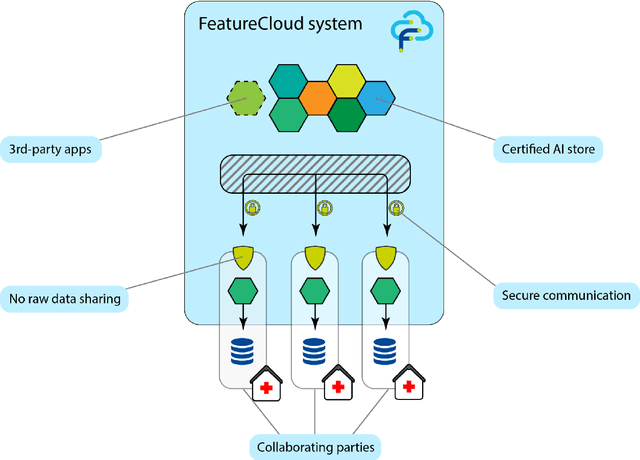
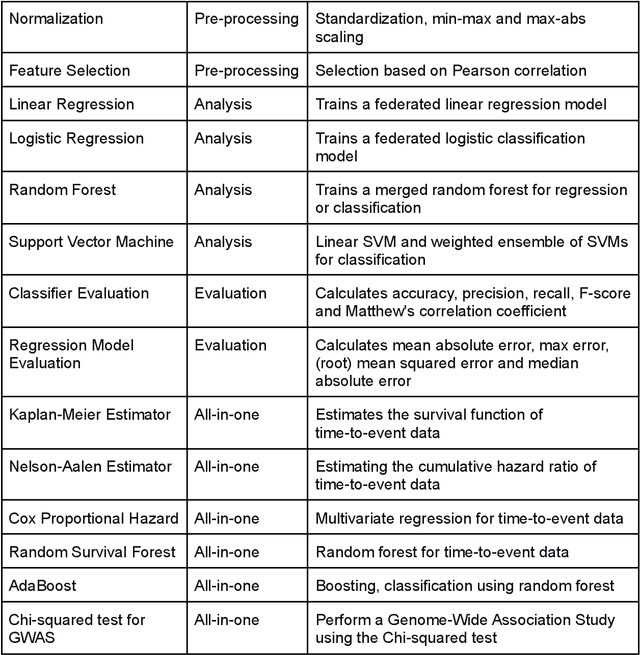
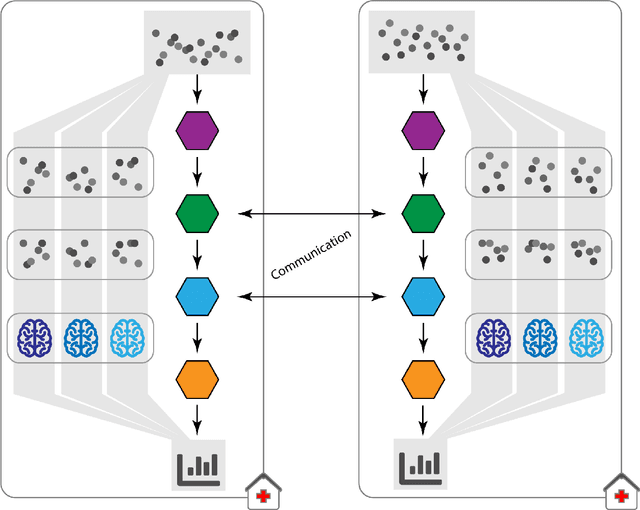
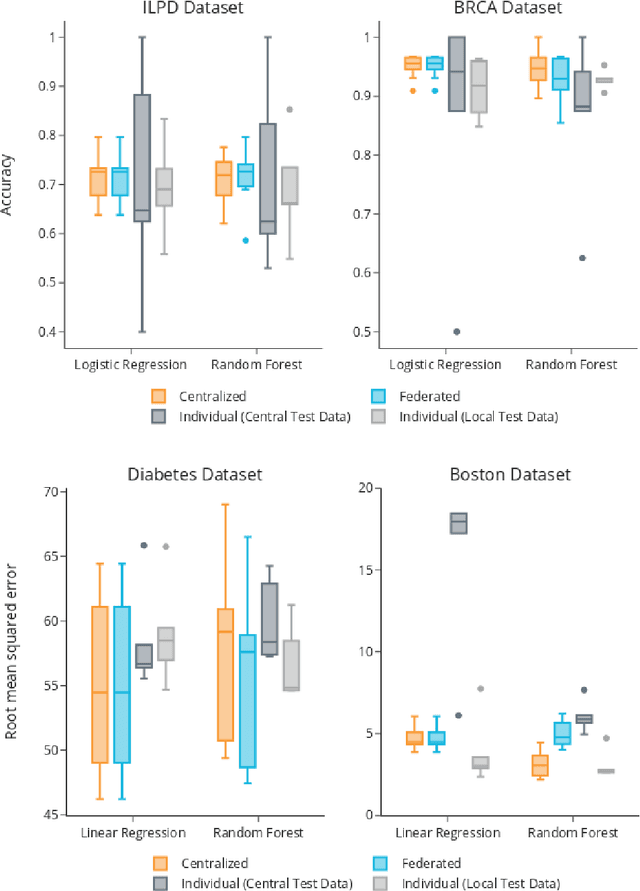
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
Privacy-preserving Artificial Intelligence Techniques in Biomedicine
Jul 22, 2020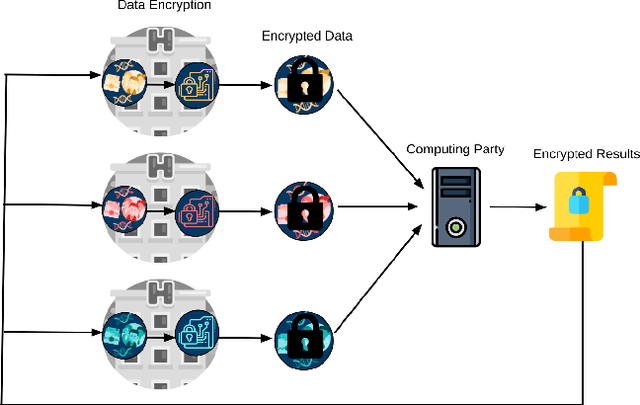
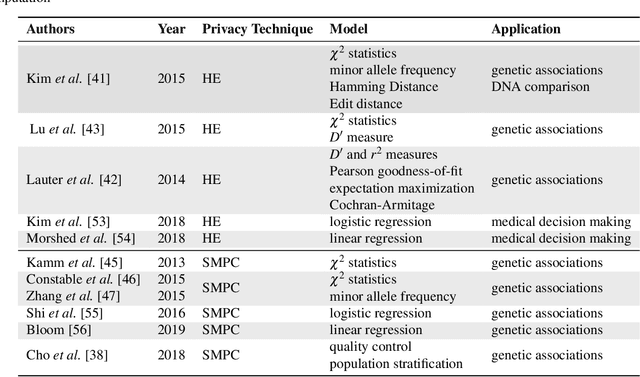
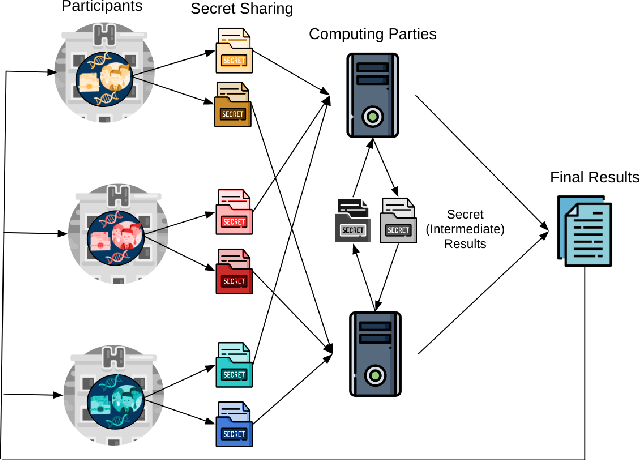
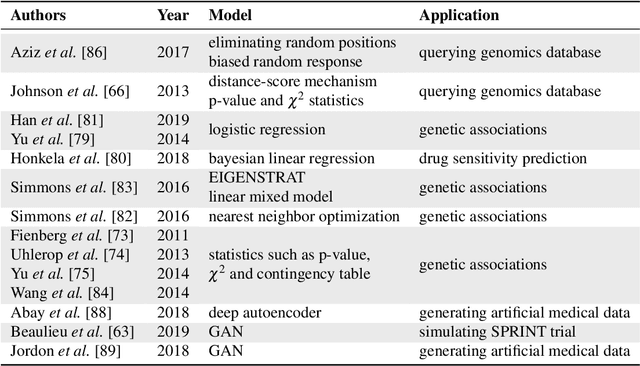
Abstract:Artificial intelligence (AI) has been successfully applied in numerous scientific domains including biomedicine and healthcare. Here, it has led to several breakthroughs ranging from clinical decision support systems, image analysis to whole genome sequencing. However, training an AI model on sensitive data raises also concerns about the privacy of individual participants. Adversary AIs, for example, can abuse even summary statistics of a study to determine the presence or absence of an individual in a given dataset. This has resulted in increasing restrictions to access biomedical data, which in turn is detrimental for collaborative research and impedes scientific progress. Hence there has been an explosive growth in efforts to harness the power of AI for learning from sensitive data while protecting patients' privacy. This paper provides a structured overview of recent advances in privacy-preserving AI techniques in biomedicine. It places the most important state-of-the-art approaches within a unified taxonomy, and discusses their strengths, limitations, and open problems.
The Virtual Doctor: An Interactive Artificial Intelligence based on Deep Learning for Non-Invasive Prediction of Diabetes
Mar 09, 2019



Abstract:Artificial intelligence (AI) will pave the way to a new era in medicine. However, currently available AI systems do not interact with a patient, e.g., for anamnesis, and thus are only used by the physicians for predictions in diagnosis or prognosis. However, these systems are widely used, e.g., in diabetes or cancer prediction. In the current study, we developed an AI that is able to interact with a patient (virtual doctor) by using a speech recognition and speech synthesis system and thus can autonomously interact with the patient, which is particularly important for, e.g., rural areas, where the availability of primary medical care is strongly limited by low population densities. As a proof-of-concept, the system is able to predict type 2 diabetes mellitus (T2DM) based on non-invasive sensors and deep neural networks. Moreover, the system provides an easy-to-interpret probability estimation for T2DM for a given patient. Besides the development of the AI, we further analyzed the acceptance of young people for AI in healthcare to estimate the impact of such system in the future.
FRI - Feature Relevance Intervals for Interpretable and Interactive Data Exploration
Mar 02, 2019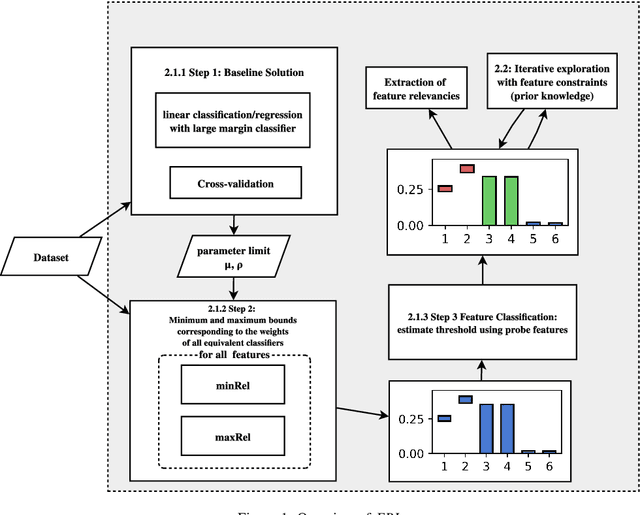

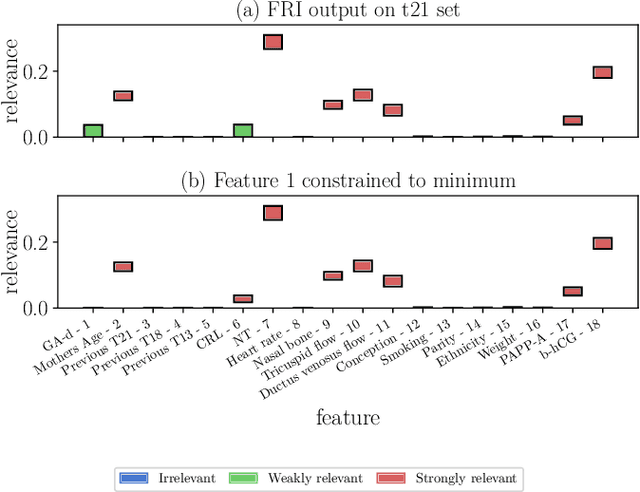
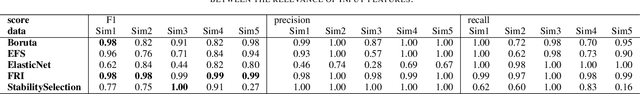
Abstract:Most existing feature selection methods are insufficient for analytic purposes as soon as high dimensional data or redundant sensor signals are dealt with since features can be selected due to spurious effects or correlations rather than causal effects. To support the finding of causal features in biomedical experiments, we hereby present FRI, an open source Python library that can be used to identify all-relevant variables in linear classification and (ordinal) regression problems. Using the recently proposed feature relevance method, FRI is able to provide the base for further general experimentation or in specific can facilitate the search for alternative biomarkers. It can be used in an interactive context, by providing model manipulation and visualization methods, or in a batch process as a filter method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge