Lukas Pfannschmidt
Sequential Feature Classification in the Context of Redundancies
Apr 15, 2020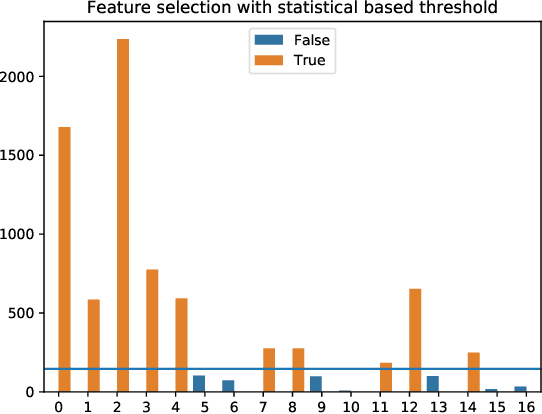
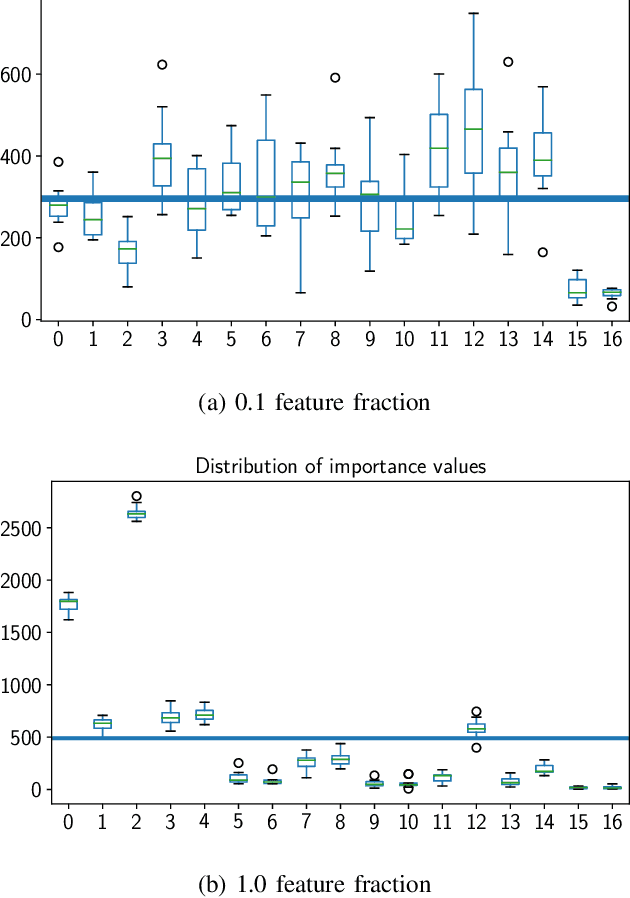

Abstract:The problem of all-relevant feature selection is concerned with finding a relevant feature set with preserved redundancies. There exist several approximations to solve this problem but only one could give a distinction between strong and weak relevance. This approach was limited to the case of linear problems. In this work, we present a new solution for this distinction in the non-linear case through the use of random forest models and statistical methods.
Feature Relevance Determination for Ordinal Regression in the Context of Feature Redundancies and Privileged Information
Dec 10, 2019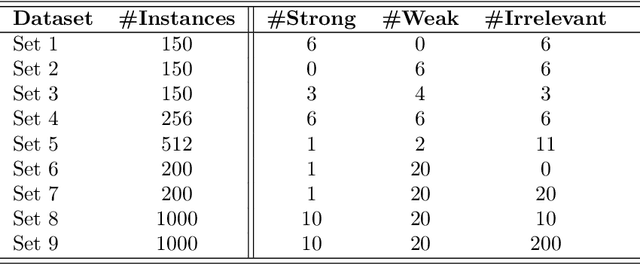
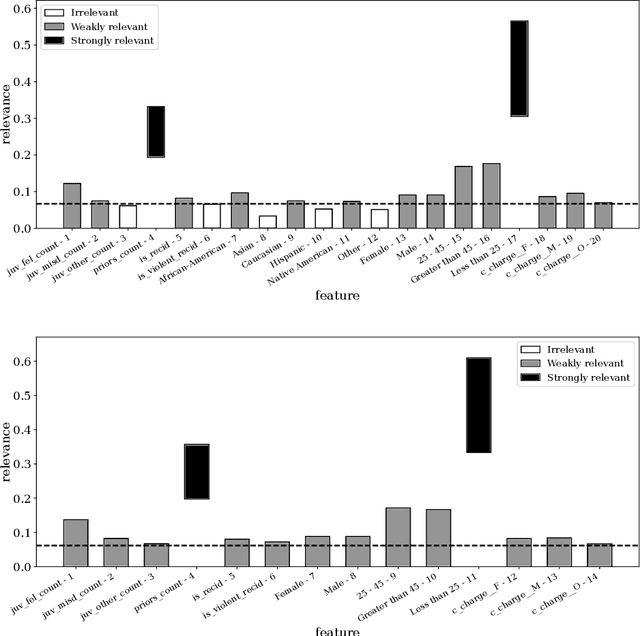
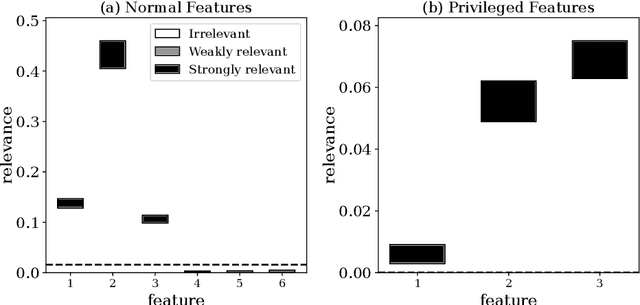
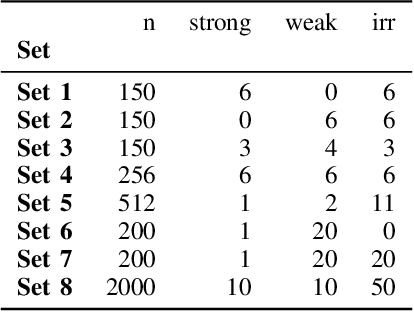
Abstract:Advances in machine learning technologies have led to increasingly powerful models in particular in the context of big data. Yet, many application scenarios demand for robustly interpretable models rather than optimum model accuracy; as an example, this is the case if potential biomarkers or causal factors should be discovered based on a set of given measurements. In this contribution, we focus on feature selection paradigms, which enable us to uncover relevant factors of a given regularity based on a sparse model. We focus on the important specific setting of linear ordinal regression, i.e.\ data have to be ranked into one of a finite number of ordered categories by a linear projection. Unlike previous work, we consider the case that features are potentially redundant, such that no unique minimum set of relevant features exists. We aim for an identification of all strongly and all weakly relevant features as well as their type of relevance (strong or weak); we achieve this goal by determining feature relevance bounds, which correspond to the minimum and maximum feature relevance, respectively, if searched over all equivalent models. In addition, we discuss how this setting enables us to substitute some of the features, e.g.\ due to their semantics, and how to extend the framework of feature relevance intervals to the setting of privileged information, i.e.\ potentially relevant information is available for training purposes only, but cannot be used for the prediction itself.
FRI - Feature Relevance Intervals for Interpretable and Interactive Data Exploration
Mar 02, 2019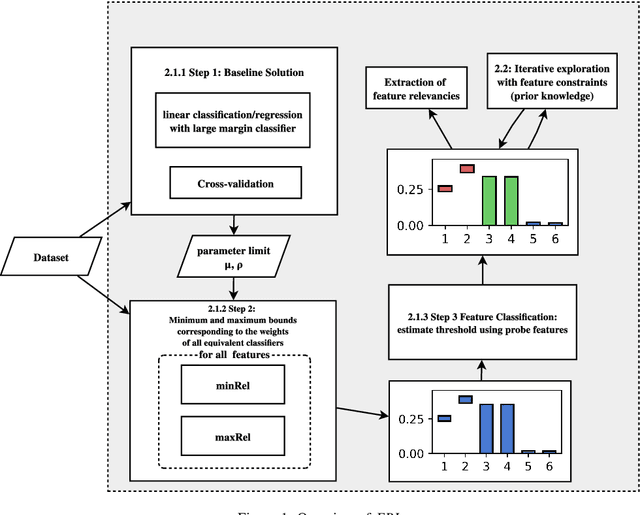

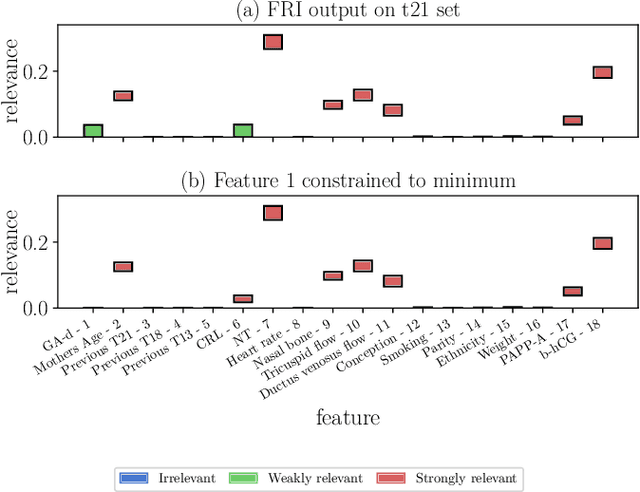
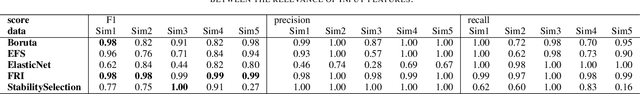
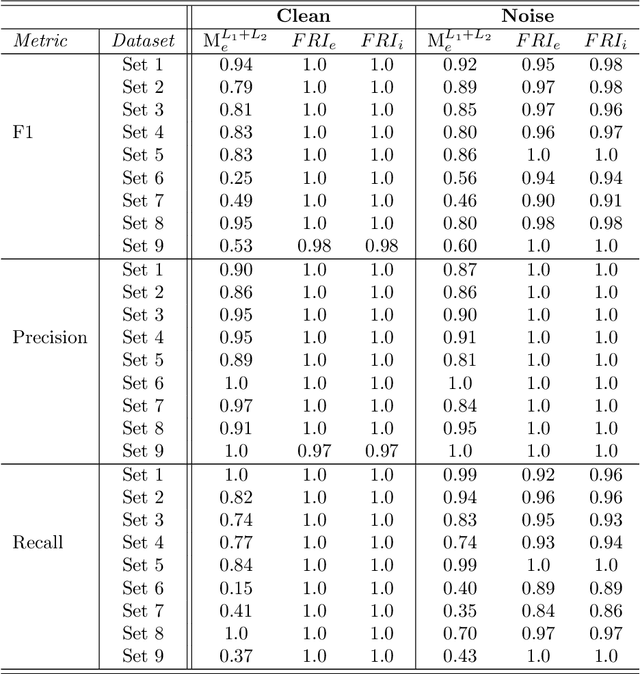
Abstract:Most existing feature selection methods are insufficient for analytic purposes as soon as high dimensional data or redundant sensor signals are dealt with since features can be selected due to spurious effects or correlations rather than causal effects. To support the finding of causal features in biomedical experiments, we hereby present FRI, an open source Python library that can be used to identify all-relevant variables in linear classification and (ordinal) regression problems. Using the recently proposed feature relevance method, FRI is able to provide the base for further general experimentation or in specific can facilitate the search for alternative biomarkers. It can be used in an interactive context, by providing model manipulation and visualization methods, or in a batch process as a filter method.
Feature Relevance Bounds for Ordinal Regression
Feb 20, 2019
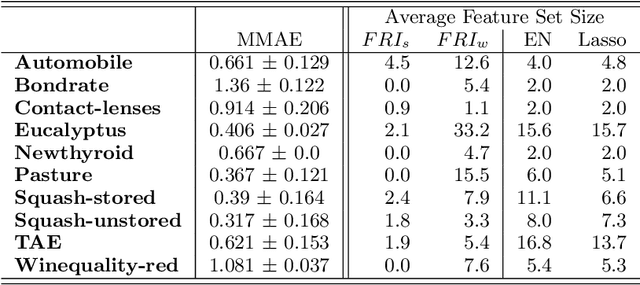
Abstract:The increasing occurrence of ordinal data, mainly sociodemographic, led to a renewed research interest in ordinal regression, i.e. the prediction of ordered classes. Besides model accuracy, the interpretation of these models itself is of high relevance, and existing approaches therefore enforce e.g. model sparsity. For high dimensional or highly correlated data, however, this might be misleading due to strong variable dependencies. In this contribution, we aim for an identification of feature relevance bounds which - besides identifying all relevant features - explicitly differentiates between strongly and weakly relevant features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge