Daoze Zhang
MOON Embedding: Multimodal Representation Learning for E-commerce Search Advertising
Nov 18, 2025Abstract:We introduce MOON, our comprehensive set of sustainable iterative practices for multimodal representation learning for e-commerce applications. MOON has already been fully deployed across all stages of Taobao search advertising system, including retrieval, relevance, ranking, and so on. The performance gains are particularly significant on click-through rate (CTR) prediction task, which achieves an overall +20.00% online CTR improvement. Over the past three years, this project has delivered the largest improvement on CTR prediction task and undergone five full-scale iterations. Throughout the exploration and iteration of our MOON, we have accumulated valuable insights and practical experience that we believe will benefit the research community. MOON contains a three-stage training paradigm of "Pretraining, Post-training, and Application", allowing effective integration of multimodal representations with downstream tasks. Notably, to bridge the misalignment between the objectives of multimodal representation learning and downstream training, we define the exchange rate to quantify how effectively improvements in an intermediate metric can translate into downstream gains. Through this analysis, we identify the image-based search recall as a critical intermediate metric guiding the optimization of multimodal models. Over three years and five iterations, MOON has evolved along four critical dimensions: data processing, training strategy, model architecture, and downstream application. The lessons and insights gained through the iterative improvements will also be shared. As part of our exploration into scaling effects in the e-commerce field, we further conduct a systematic study of the scaling laws governing multimodal representation learning, examining multiple factors such as the number of training tokens, negative samples, and the length of user behavior sequences.
MOON2.0: Dynamic Modality-balanced Multimodal Representation Learning for E-commerce Product Understanding
Nov 16, 2025Abstract:The rapid growth of e-commerce calls for multimodal models that comprehend rich visual and textual product information. Although recent multimodal large language models (MLLMs) for product understanding exhibit strong capability in representation learning for e-commerce, they still face three challenges: (i) the modality imbalance induced by modality mixed training; (ii) underutilization of the intrinsic alignment relationships among visual and textual information within a product; and (iii) limited handling of noise in e-commerce multimodal data. To address these, we propose MOON2.0, a dynamic modality-balanced multimodal representation learning framework for e-commerce product understanding. MOON2.0 comprises: (1) a Modality-driven Mixture-of-Experts (MoE) module that adaptively processes input samples by their modality composition, enabling Multimodal Joint Learning to mitigate the modality imbalance; (2) a Dual-level Alignment method to better leverage semantic alignment properties inside individual products; and (3) an MLLM-based Image-text Co-augmentation strategy that integrates textual enrichment with visual expansion, coupled with Dynamic Sample Filtering to improve training data quality. We further introduce MBE2.0, a co-augmented multimodal representation benchmark for e-commerce representation learning and evaluation. Experiments show that MOON2.0 delivers state-of-the-art zero-shot performance on MBE2.0 and multiple public datasets. Furthermore, attention-based heatmap visualization provides qualitative evidence of improved multimodal alignment of MOON2.0.
PD$^3$: A Project Duplication Detection Framework via Adapted Multi-Agent Debate
May 23, 2025Abstract:Project duplication detection is critical for project quality assessment, as it improves resource utilization efficiency by preventing investing in newly proposed project that have already been studied. It requires the ability to understand high-level semantics and generate constructive and valuable feedback. Existing detection methods rely on basic word- or sentence-level comparison or solely apply large language models, lacking valuable insights for experts and in-depth comprehension of project content and review criteria. To tackle this issue, we propose PD$^3$, a Project Duplication Detection framework via adapted multi-agent Debate. Inspired by real-world expert debates, it employs a fair competition format to guide multi-agent debate to retrieve relevant projects. For feedback, it incorporates both qualitative and quantitative analysis to improve its practicality. Over 800 real-world power project data spanning more than 20 specialized fields are used to evaluate the framework, demonstrating that our method outperforms existing approaches by 7.43% and 8.00% in two downstream tasks. Furthermore, we establish an online platform, Review Dingdang, to assist power experts, saving 5.73 million USD in initial detection on more than 100 newly proposed projects.
Re$^2$: A Consistency-ensured Dataset for Full-stage Peer Review and Multi-turn Rebuttal Discussions
May 12, 2025Abstract:Peer review is a critical component of scientific progress in the fields like AI, but the rapid increase in submission volume has strained the reviewing system, which inevitably leads to reviewer shortages and declines review quality. Besides the growing research popularity, another key factor in this overload is the repeated resubmission of substandard manuscripts, largely due to the lack of effective tools for authors to self-evaluate their work before submission. Large Language Models (LLMs) show great promise in assisting both authors and reviewers, and their performance is fundamentally limited by the quality of the peer review data. However, existing peer review datasets face three major limitations: (1) limited data diversity, (2) inconsistent and low-quality data due to the use of revised rather than initial submissions, and (3) insufficient support for tasks involving rebuttal and reviewer-author interactions. To address these challenges, we introduce the largest consistency-ensured peer review and rebuttal dataset named Re^2, which comprises 19,926 initial submissions, 70,668 review comments, and 53,818 rebuttals from 24 conferences and 21 workshops on OpenReview. Moreover, the rebuttal and discussion stage is framed as a multi-turn conversation paradigm to support both traditional static review tasks and dynamic interactive LLM assistants, providing more practical guidance for authors to refine their manuscripts and helping alleviate the growing review burden. Our data and code are available in https://anonymous.4open.science/r/ReviewBench_anon/.
Deep Learning-Powered Electrical Brain Signals Analysis: Advancing Neurological Diagnostics
Feb 24, 2025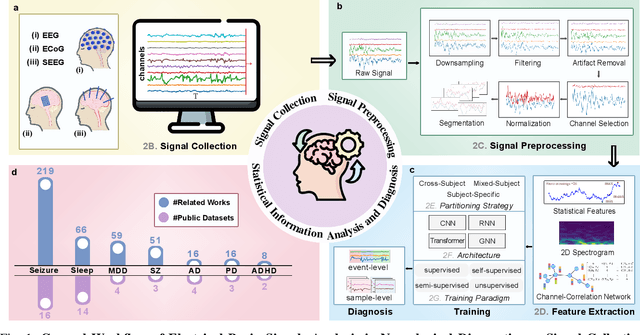
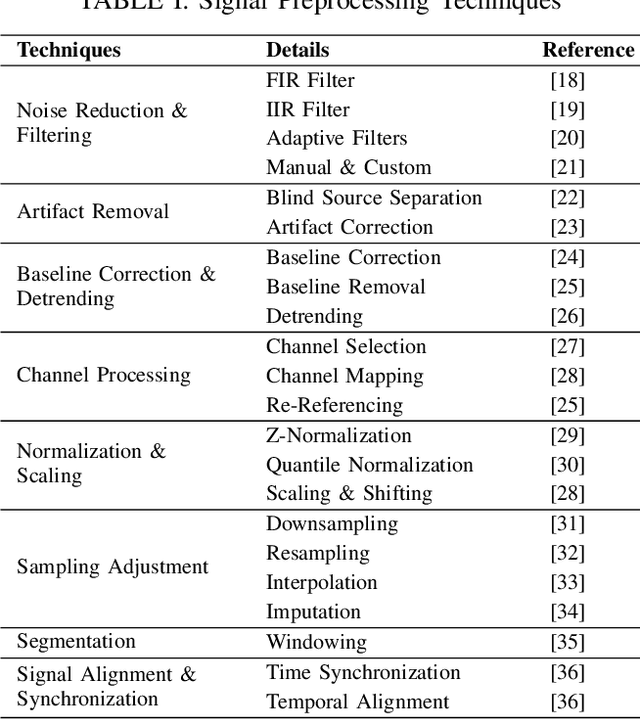
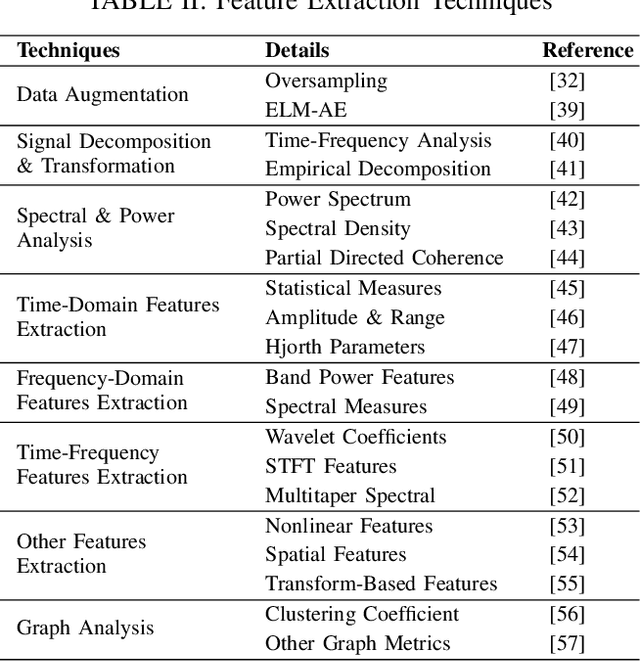
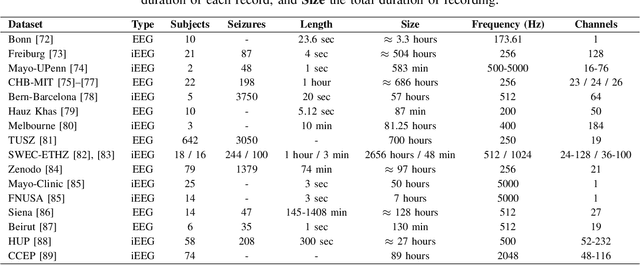
Abstract:Neurological disorders represent significant global health challenges, driving the advancement of brain signal analysis methods. Scalp electroencephalography (EEG) and intracranial electroencephalography (iEEG) are widely used to diagnose and monitor neurological conditions. However, dataset heterogeneity and task variations pose challenges in developing robust deep learning solutions. This review systematically examines recent advances in deep learning approaches for EEG/iEEG-based neurological diagnostics, focusing on applications across 7 neurological conditions using 46 datasets. We explore trends in data utilization, model design, and task-specific adaptations, highlighting the importance of pre-trained multi-task models for scalable, generalizable solutions. To advance research, we propose a standardized benchmark for evaluating models across diverse datasets to enhance reproducibility. This survey emphasizes how recent innovations can transform neurological diagnostics and enable the development of intelligent, adaptable healthcare solutions.
Minimum Tuning to Unlock Long Output from LLMs with High Quality Data as the Key
Oct 15, 2024Abstract:As large language models rapidly evolve to support longer context, there is a notable disparity in their capability to generate output at greater lengths. Recent study suggests that the primary cause for this imbalance may arise from the lack of data with long-output during alignment training. In light of this observation, attempts are made to re-align foundation models with data that fills the gap, which result in models capable of generating lengthy output when instructed. In this paper, we explore the impact of data-quality in tuning a model for long output, and the possibility of doing so from the starting points of human-aligned (instruct or chat) models. With careful data curation, we show that it possible to achieve similar performance improvement in our tuned models, with only a small fraction of training data instances and compute. In addition, we assess the generalizability of such approaches by applying our tuning-recipes to several models. our findings suggest that, while capacities for generating long output vary across different models out-of-the-box, our approach to tune them with high-quality data using lite compute, consistently yields notable improvement across all models we experimented on. We have made public our curated dataset for tuning long-writing capability, the implementations of model tuning and evaluation, as well as the fine-tuned models, all of which can be openly-accessed.
Brant-X: A Unified Physiological Signal Alignment Framework
Aug 28, 2024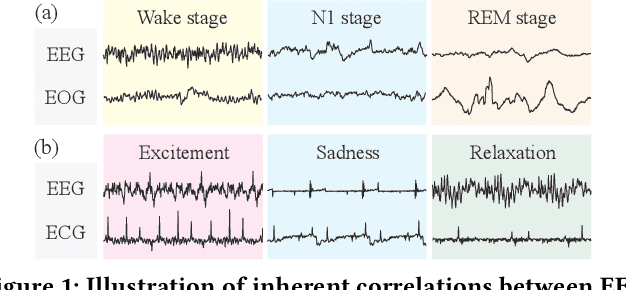
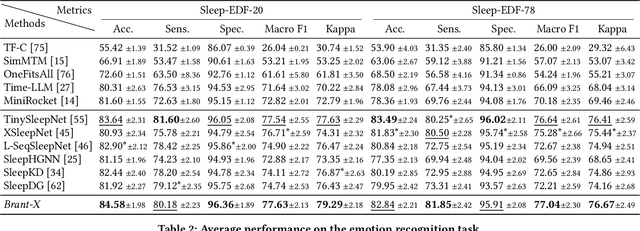
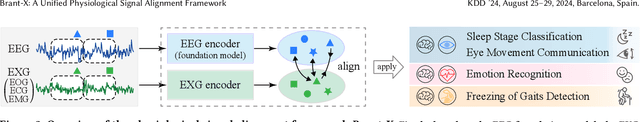
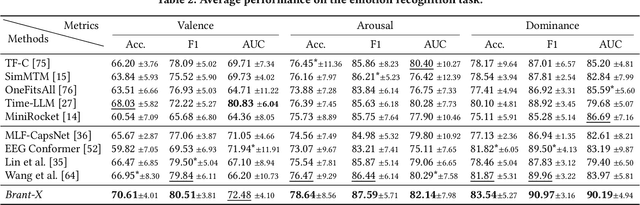
Abstract:Physiological signals serve as indispensable clues for understanding various physiological states of human bodies. Most existing works have focused on a single type of physiological signals for a range of application scenarios. However, as the body is a holistic biological system, the inherent interconnection among various physiological data should not be neglected. In particular, given the brain's role as the control center for vital activities, electroencephalogram (EEG) exhibits significant correlations with other physiological signals. Therefore, the correlation between EEG and other physiological signals holds potential to improve performance in various scenarios. Nevertheless, achieving this goal is still constrained by several challenges: the scarcity of simultaneously collected physiological data, the differences in correlations between various signals, and the correlation differences between various tasks. To address these issues, we propose a unified physiological signal alignment framework, Brant-X, to model the correlation between EEG and other signals. Our approach (1) employs the EEG foundation model to data-efficiently transfer the rich knowledge in EEG to other physiological signals, and (2) introduces the two-level alignment to fully align the semantics of EEG and other signals from different semantic scales. In the experiments, Brant-X achieves state-of-the-art performance compared with task-agnostic and task-specific baselines on various downstream tasks in diverse scenarios, including sleep stage classification, emotion recognition, freezing of gaits detection, and eye movement communication. Moreover, the analysis on the arrhythmia detection task and the visualization in case study further illustrate the effectiveness of Brant-X in the knowledge transfer from EEG to other physiological signals. The model's homepage is at https://github.com/zjunet/Brant-X/.
* Accepted by SIGKDD 2024
SWIFT:A Scalable lightWeight Infrastructure for Fine-Tuning
Aug 13, 2024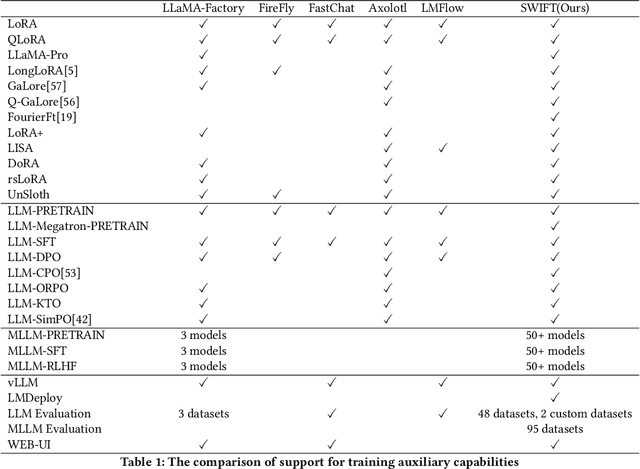
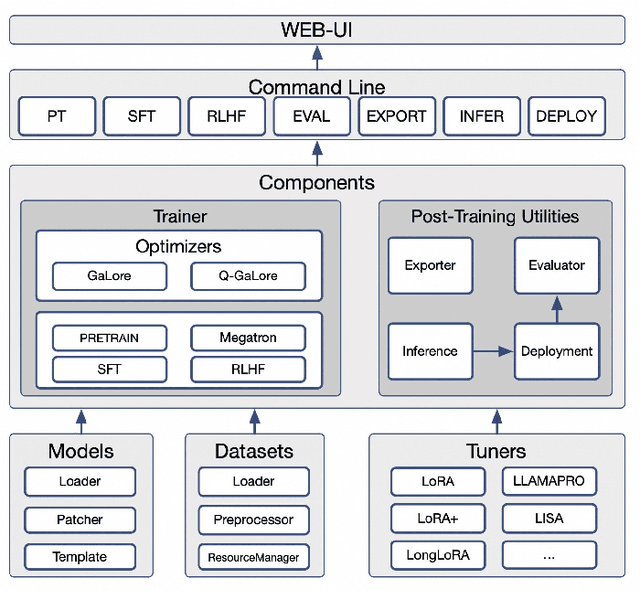
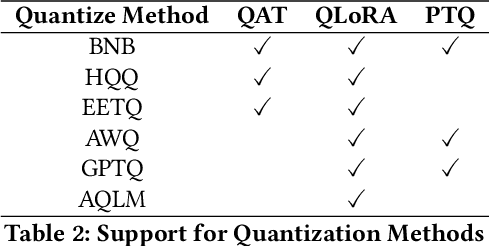
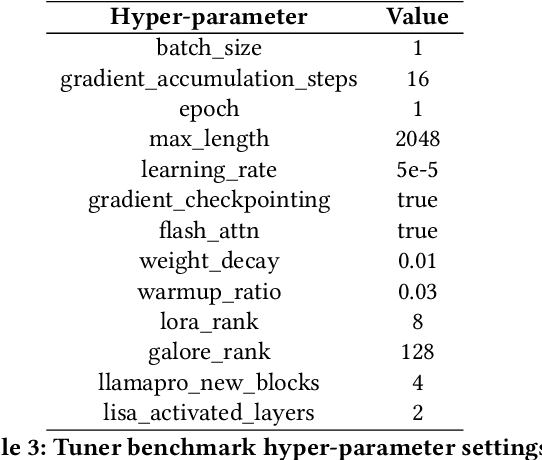
Abstract:Recent development in Large Language Models (LLMs) and Multi-modal Large Language Models (MLLMs) have leverage Attention-based Transformer architectures and achieved superior performance and generalization capabilities. They have since covered extensive areas of traditional learning tasks. For instance, text-based tasks such as text-classification and sequence-labeling, as well as multi-modal tasks like Visual Question Answering (VQA) and Optical Character Recognition (OCR), which were previously addressed using different models, can now be tackled based on one foundation model. Consequently, the training and lightweight fine-tuning of LLMs and MLLMs, especially those based on Transformer architecture, has become particularly important. In recognition of these overwhelming needs, we develop SWIFT, a customizable one-stop infrastructure for large models. With support of over $300+$ LLMs and $50+$ MLLMs, SWIFT stands as the open-source framework that provide the \textit{most comprehensive support} for fine-tuning large models. In particular, it is the first training framework that provides systematic support for MLLMs. In addition to the core functionalities of fine-tuning, SWIFT also integrates post-training processes such as inference, evaluation, and model quantization, to facilitate fast adoptions of large models in various application scenarios. With a systematic integration of various training techniques, SWIFT offers helpful utilities such as benchmark comparisons among different training techniques for large models. For fine-tuning models specialized in agent framework, we show that notable improvements on the ToolBench leader-board can be achieved by training with customized dataset on SWIFT, with an increase of 5.2%-21.8% in the Act.EM metric over various baseline models, a reduction in hallucination by 1.6%-14.1%, and an average performance improvement of 8%-17%.
Brant-2: Foundation Model for Brain Signals
Feb 22, 2024Abstract:Foundational models benefit from pre-training on large amounts of unlabeled data and enable strong performance in a wide variety of applications with a small amount of labeled data. Such models can be particularly effective in analyzing brain signals, as this field encompasses numerous application scenarios, and it is costly to perform large-scale annotation. In this work, we present the largest foundation model in brain signals, Brant-2. Compared to Brant, a foundation model designed for intracranial neural signals, Brant-2 not only exhibits robustness towards data variations and modeling scales but also can be applied to a broader range of brain neural data. By experimenting on an extensive range of tasks, we demonstrate that Brant-2 is adaptive to various application scenarios in brain signals. Further analyses reveal the scalability of the Brant-2, validate each component's effectiveness, and showcase our model's ability to maintain performance in scenarios with scarce labels. The source code and pre-trained weights are available at: https://github.com/yzz673/Brant-2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge