Zhizhang Yuan
Assembling the Mind's Mosaic: Towards EEG Semantic Intent Decoding
Jan 28, 2026Abstract:Enabling natural communication through brain-computer interfaces (BCIs) remains one of the most profound challenges in neuroscience and neurotechnology. While existing frameworks offer partial solutions, they are constrained by oversimplified semantic representations and a lack of interpretability. To overcome these limitations, we introduce Semantic Intent Decoding (SID), a novel framework that translates neural activity into natural language by modeling meaning as a flexible set of compositional semantic units. SID is built on three core principles: semantic compositionality, continuity and expandability of semantic space, and fidelity in reconstruction. We present BrainMosaic, a deep learning architecture implementing SID. BrainMosaic decodes multiple semantic units from EEG/SEEG signals using set matching and then reconstructs coherent sentences through semantic-guided reconstruction. This approach moves beyond traditional pipelines that rely on fixed-class classification or unconstrained generation, enabling a more interpretable and expressive communication paradigm. Extensive experiments on multilingual EEG and clinical SEEG datasets demonstrate that SID and BrainMosaic offer substantial advantages over existing frameworks, paving the way for natural and effective BCI-mediated communication.
NeuroSketch: An Effective Framework for Neural Decoding via Systematic Architectural Optimization
Dec 10, 2025Abstract:Neural decoding, a critical component of Brain-Computer Interface (BCI), has recently attracted increasing research interest. Previous research has focused on leveraging signal processing and deep learning methods to enhance neural decoding performance. However, the in-depth exploration of model architectures remains underexplored, despite its proven effectiveness in other tasks such as energy forecasting and image classification. In this study, we propose NeuroSketch, an effective framework for neural decoding via systematic architecture optimization. Starting with the basic architecture study, we find that CNN-2D outperforms other architectures in neural decoding tasks and explore its effectiveness from temporal and spatial perspectives. Building on this, we optimize the architecture from macro- to micro-level, achieving improvements in performance at each step. The exploration process and model validations take over 5,000 experiments spanning three distinct modalities (visual, auditory, and speech), three types of brain signals (EEG, SEEG, and ECoG), and eight diverse decoding tasks. Experimental results indicate that NeuroSketch achieves state-of-the-art (SOTA) performance across all evaluated datasets, positioning it as a powerful tool for neural decoding. Our code and scripts are available at https://github.com/Galaxy-Dawn/NeuroSketch.
Deep Learning-Powered Electrical Brain Signals Analysis: Advancing Neurological Diagnostics
Feb 24, 2025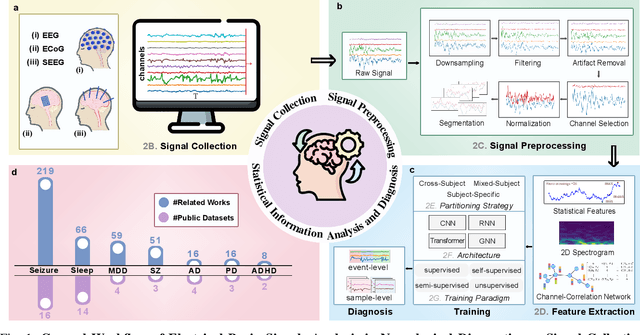
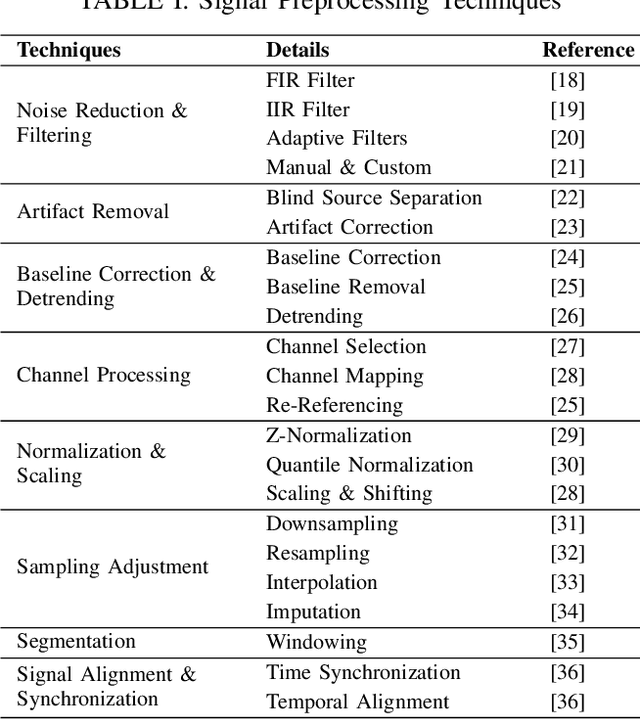
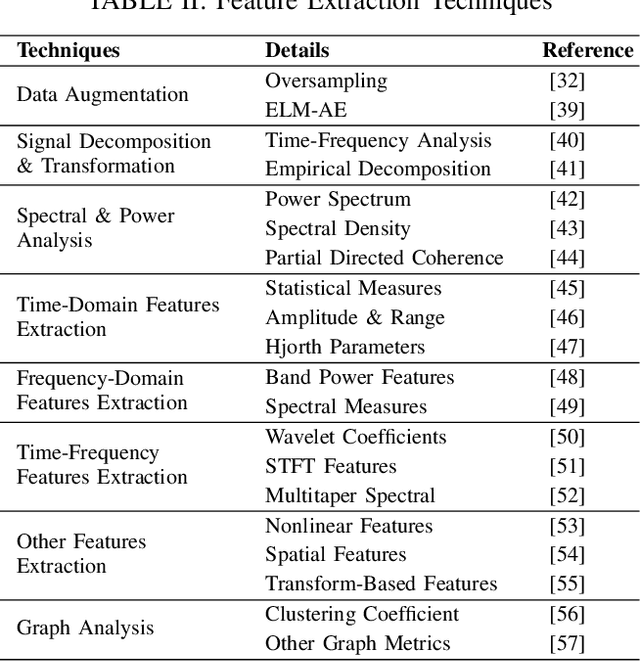
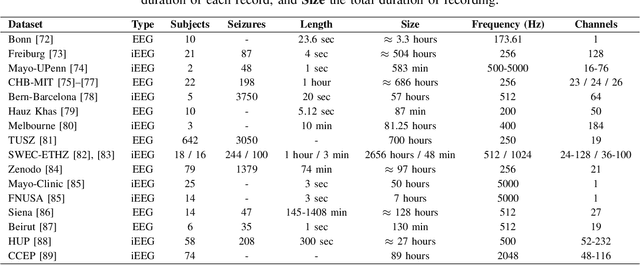
Abstract:Neurological disorders represent significant global health challenges, driving the advancement of brain signal analysis methods. Scalp electroencephalography (EEG) and intracranial electroencephalography (iEEG) are widely used to diagnose and monitor neurological conditions. However, dataset heterogeneity and task variations pose challenges in developing robust deep learning solutions. This review systematically examines recent advances in deep learning approaches for EEG/iEEG-based neurological diagnostics, focusing on applications across 7 neurological conditions using 46 datasets. We explore trends in data utilization, model design, and task-specific adaptations, highlighting the importance of pre-trained multi-task models for scalable, generalizable solutions. To advance research, we propose a standardized benchmark for evaluating models across diverse datasets to enhance reproducibility. This survey emphasizes how recent innovations can transform neurological diagnostics and enable the development of intelligent, adaptable healthcare solutions.
Brant-X: A Unified Physiological Signal Alignment Framework
Aug 28, 2024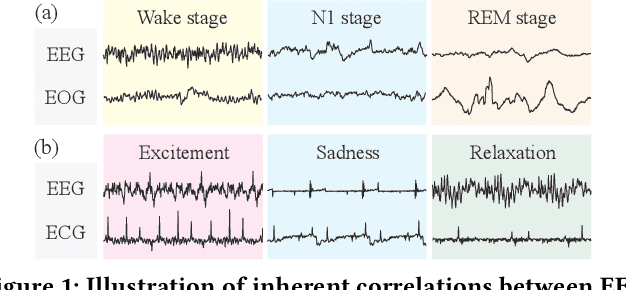
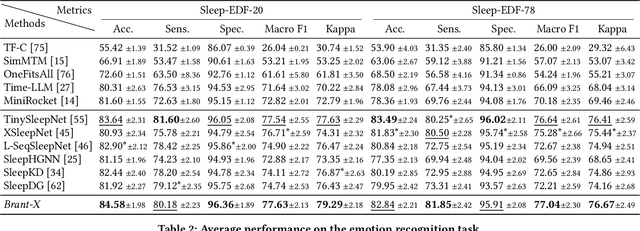
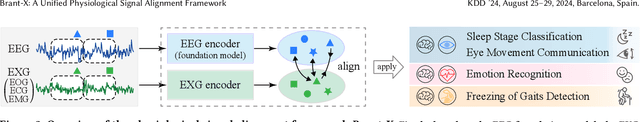
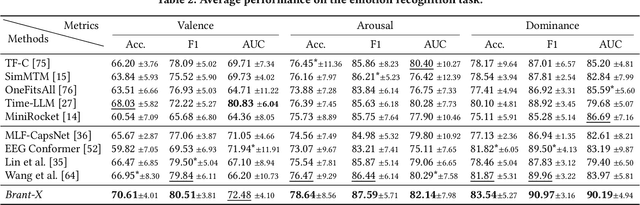
Abstract:Physiological signals serve as indispensable clues for understanding various physiological states of human bodies. Most existing works have focused on a single type of physiological signals for a range of application scenarios. However, as the body is a holistic biological system, the inherent interconnection among various physiological data should not be neglected. In particular, given the brain's role as the control center for vital activities, electroencephalogram (EEG) exhibits significant correlations with other physiological signals. Therefore, the correlation between EEG and other physiological signals holds potential to improve performance in various scenarios. Nevertheless, achieving this goal is still constrained by several challenges: the scarcity of simultaneously collected physiological data, the differences in correlations between various signals, and the correlation differences between various tasks. To address these issues, we propose a unified physiological signal alignment framework, Brant-X, to model the correlation between EEG and other signals. Our approach (1) employs the EEG foundation model to data-efficiently transfer the rich knowledge in EEG to other physiological signals, and (2) introduces the two-level alignment to fully align the semantics of EEG and other signals from different semantic scales. In the experiments, Brant-X achieves state-of-the-art performance compared with task-agnostic and task-specific baselines on various downstream tasks in diverse scenarios, including sleep stage classification, emotion recognition, freezing of gaits detection, and eye movement communication. Moreover, the analysis on the arrhythmia detection task and the visualization in case study further illustrate the effectiveness of Brant-X in the knowledge transfer from EEG to other physiological signals. The model's homepage is at https://github.com/zjunet/Brant-X/.
* Accepted by SIGKDD 2024
Brant-2: Foundation Model for Brain Signals
Feb 22, 2024Abstract:Foundational models benefit from pre-training on large amounts of unlabeled data and enable strong performance in a wide variety of applications with a small amount of labeled data. Such models can be particularly effective in analyzing brain signals, as this field encompasses numerous application scenarios, and it is costly to perform large-scale annotation. In this work, we present the largest foundation model in brain signals, Brant-2. Compared to Brant, a foundation model designed for intracranial neural signals, Brant-2 not only exhibits robustness towards data variations and modeling scales but also can be applied to a broader range of brain neural data. By experimenting on an extensive range of tasks, we demonstrate that Brant-2 is adaptive to various application scenarios in brain signals. Further analyses reveal the scalability of the Brant-2, validate each component's effectiveness, and showcase our model's ability to maintain performance in scenarios with scarce labels. The source code and pre-trained weights are available at: https://github.com/yzz673/Brant-2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge