Craig Meyer
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021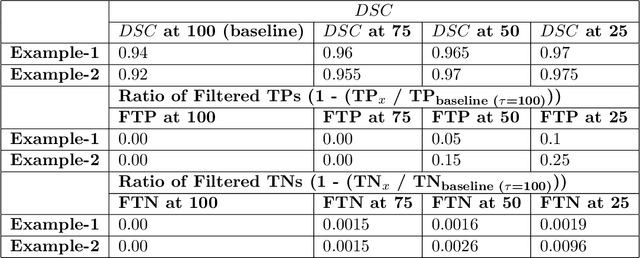
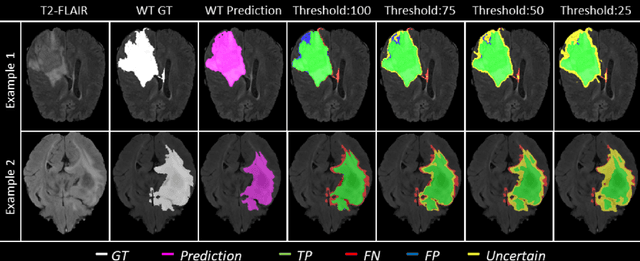
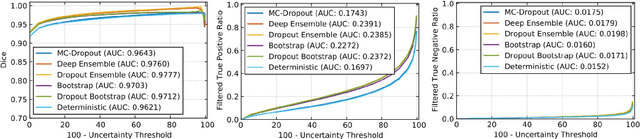
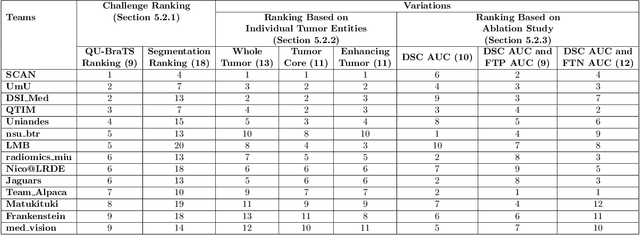
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Deep Learning methods for automatic evaluation of delayed enhancement-MRI. The results of the EMIDEC challenge
Aug 10, 2021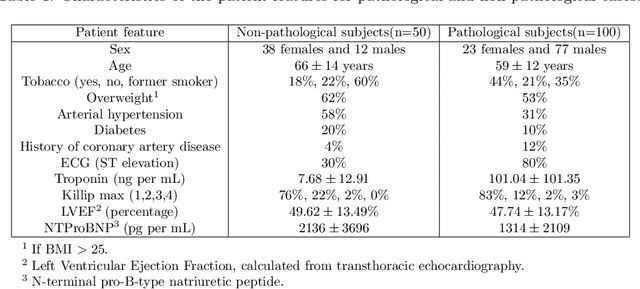
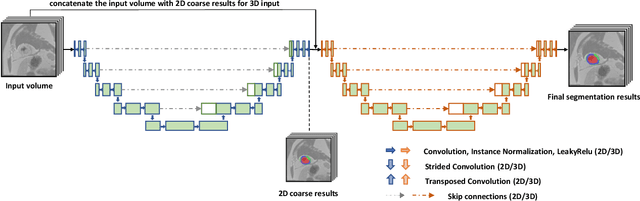
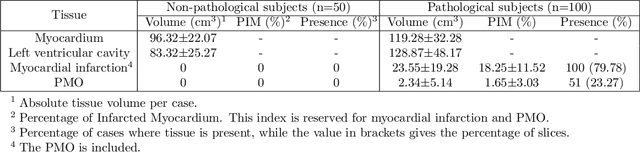
Abstract:A key factor for assessing the state of the heart after myocardial infarction (MI) is to measure whether the myocardium segment is viable after reperfusion or revascularization therapy. Delayed enhancement-MRI or DE-MRI, which is performed several minutes after injection of the contrast agent, provides high contrast between viable and nonviable myocardium and is therefore a method of choice to evaluate the extent of MI. To automatically assess myocardial status, the results of the EMIDEC challenge that focused on this task are presented in this paper. The challenge's main objectives were twofold. First, to evaluate if deep learning methods can distinguish between normal and pathological cases. Second, to automatically calculate the extent of myocardial infarction. The publicly available database consists of 150 exams divided into 50 cases with normal MRI after injection of a contrast agent and 100 cases with myocardial infarction (and then with a hyperenhanced area on DE-MRI), whatever their inclusion in the cardiac emergency department. Along with MRI, clinical characteristics are also provided. The obtained results issued from several works show that the automatic classification of an exam is a reachable task (the best method providing an accuracy of 0.92), and the automatic segmentation of the myocardium is possible. However, the segmentation of the diseased area needs to be improved, mainly due to the small size of these areas and the lack of contrast with the surrounding structures.
Prostate Segmentation from 3D MRI Using a Two-Stage Model and Variable-Input Based Uncertainty Measure
Mar 06, 2019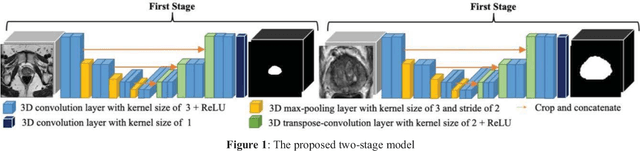
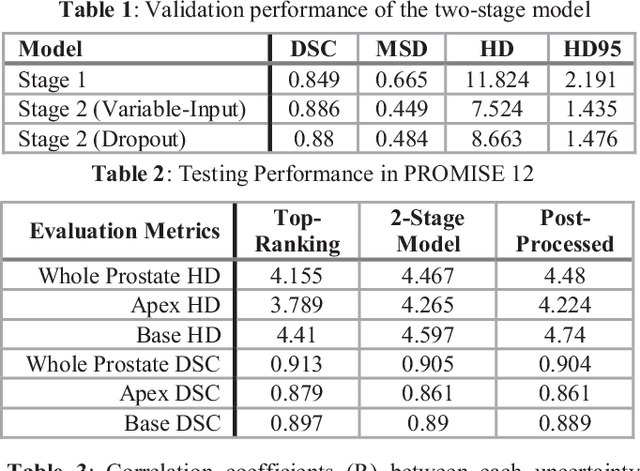
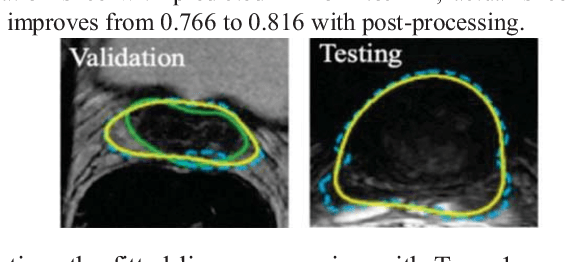
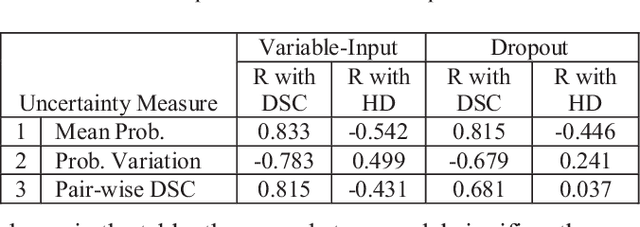
Abstract:This paper proposes a two-stage segmentation model, variable-input based uncertainty measures and an uncertainty-guided post-processing method for prostate segmentation on 3D magnetic resonance images (MRI). The two-stage model was based on 3D dilated U-Nets with the first stage to localize the prostate and the second stage to obtain an accurate segmentation from cropped images. For data augmentation, we proposed the variable-input method which crops the region of interest with additional random variations. Similar to other deep learning models, the proposed model also faced the challenge of suboptimal performance in certain testing cases due to varied training and testing image characteristics. Therefore, it is valuable to evaluate the confidence and performance of the network using uncertainty measures, which are often calculated from the probability maps or their standard deviations with multiple model outputs for the same testing case. However, few studies have quantitatively compared different methods of uncertainty calculation. Furthermore, unlike the commonly used Bayesian dropout during testing, we developed uncertainty measures based on the variable input images at the second stage and evaluated its performance by calculating the correlation with ground-truth-based performance metrics, such as Dice score. For performance estimation, we predicted Dice scores and Hausdorff distance with the most correlated uncertainty measure. For post-processing, we performed Gaussian filter on the underperformed slices to improve segmentation quality. Using PROMISE-12 data, we demonstrated the robustness of the two-stage model and showed high correlation of the proposed variable-input based uncertainty measures with GT-based performance. The uncertainty-guided post-processing method significantly improved label smoothness.
Brain Tumor Segmentation using an Ensemble of 3D U-Nets and Overall Survival Prediction using Radiomic Features
Dec 03, 2018
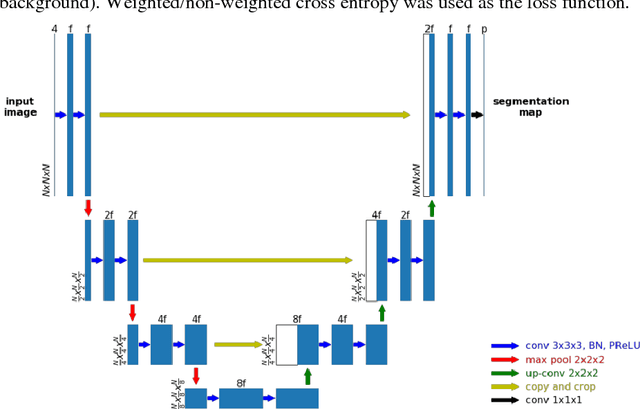
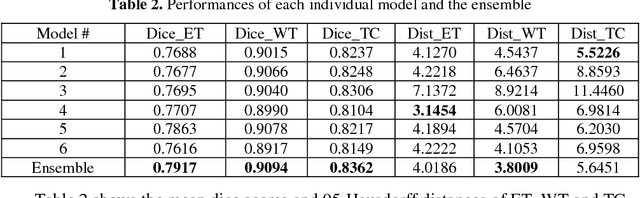
Abstract:Accurate segmentation of different sub-regions of gliomas including peritumoral edema, necrotic core, enhancing and non-enhancing tumor core from multimodal MRI scans has important clinical relevance in diagnosis, prognosis and treatment of brain tumors. However, due to the highly heterogeneous appearance and shape, segmentation of the sub-regions is very challenging. Recent development using deep learning models has proved its effectiveness in the past several brain segmentation challenges as well as other semantic and medical image segmentation problems. Most models in brain tumor segmentation use a 2D/3D patch to predict the class label for the center voxel and variant patch sizes and scales are used to improve the model performance. However, it has low computation efficiency and also has limited receptive field. U-Net is a widely used network structure for end-to-end segmentation and can be used on the entire image or extracted patches to provide classification labels over the entire input voxels so that it is more efficient and expect to yield better performance with larger input size. Furthermore, instead of picking the best network structure, an ensemble of multiple models, trained on different dataset or different hyper-parameters, can generally improve the segmentation performance. In this study we propose to use an ensemble of 3D U-Nets with different hyper-parameters for brain tumor segmentation. Preliminary results showed effectiveness of this model. In addition, we developed a linear model for survival prediction using extracted imaging and non-imaging features, which, despite the simplicity, can effectively reduce overfitting and regression errors.
A Self-Adaptive Network For Multiple Sclerosis Lesion Segmentation From Multi-Contrast MRI With Various Imaging Protocols
Nov 19, 2018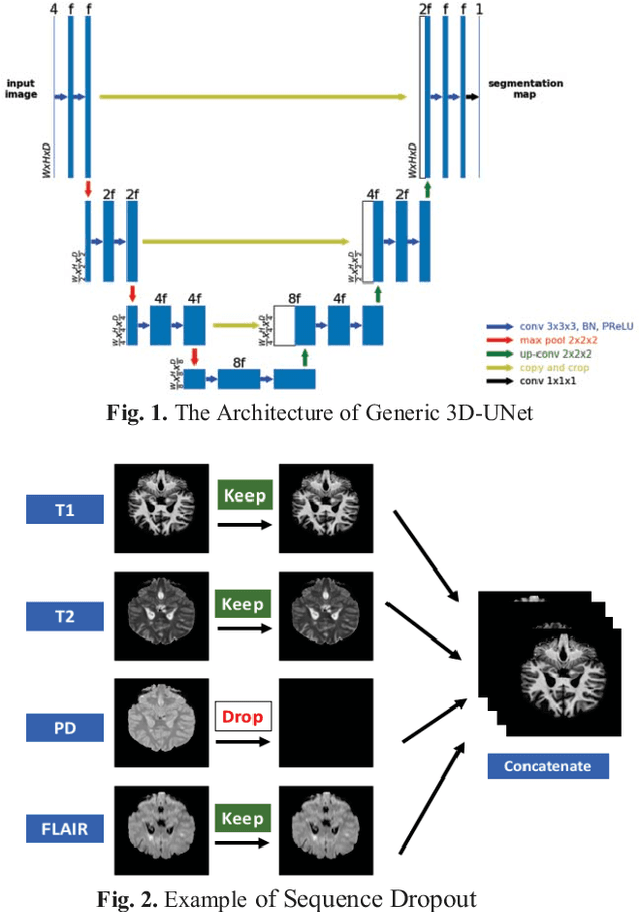
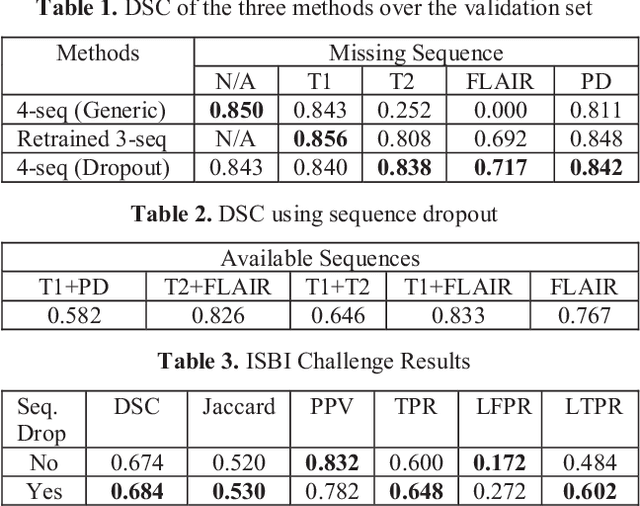
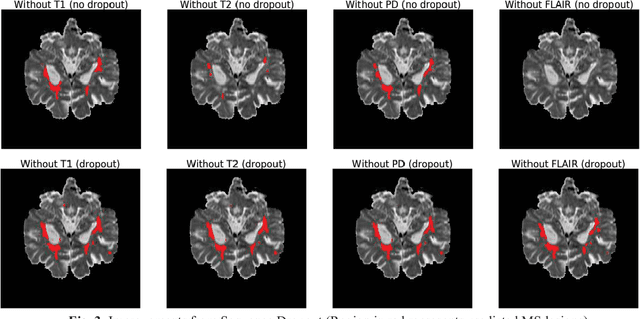
Abstract:Deep neural networks (DNN) have shown promises in the lesion segmentation of multiple sclerosis (MS) from multicontrast MRI including T1, T2, proton density (PD) and FLAIR sequences. However, one challenge in deploying such networks into clinical practice is the variability of imaging protocols, which often differ from the training dataset as certain MRI sequences may be unavailable or unusable. Therefore, trained networks need to adapt to practical situations when imaging protocols are different in deployment. In this paper, we propose a DNN-based MS lesion segmentation framework with a novel technique called sequence dropout which can adapt to various combinations of input MRI sequences during deployment and achieve the maximal possible performance from the given input. In addition, with this framework, we studied the quantitative impact of each MRI sequence on the MS lesion segmentation task without training separate networks. Experiments were performed using the IEEE ISBI 2015 Longitudinal MS Lesion Challenge dataset and our method is currently ranked 2nd with a Dice similarity coefficient of 0.684. Furthermore, we showed our network achieved the maximal possible performance when one sequence is unavailable during deployment by comparing with separate networks trained on the corresponding input MRI sequences. In particular, we discovered T1 and PD have minor impact on segmentation performance while FLAIR is the predominant sequence. Experiments with multiple missing sequences were also performed and showed the robustness of our network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge