Huitong Pan
DynClean: Training Dynamics-based Label Cleaning for Distantly-Supervised Named Entity Recognition
Apr 06, 2025Abstract:Distantly Supervised Named Entity Recognition (DS-NER) has attracted attention due to its scalability and ability to automatically generate labeled data. However, distant annotation introduces many mislabeled instances, limiting its performance. Most of the existing work attempt to solve this problem by developing intricate models to learn from the noisy labels. An alternative approach is to attempt to clean the labeled data, thus increasing the quality of distant labels. This approach has received little attention for NER. In this paper, we propose a training dynamics-based label cleaning approach, which leverages the behavior of a model as training progresses to characterize the distantly annotated samples. We also introduce an automatic threshold estimation strategy to locate the errors in distant labels. Extensive experimental results demonstrate that: (1) models trained on our cleaned DS-NER datasets, which were refined by directly removing identified erroneous annotations, achieve significant improvements in F1-score, ranging from 3.18% to 8.95%; and (2) our method outperforms numerous advanced DS-NER approaches across four datasets.
SciER: An Entity and Relation Extraction Dataset for Datasets, Methods, and Tasks in Scientific Documents
Oct 28, 2024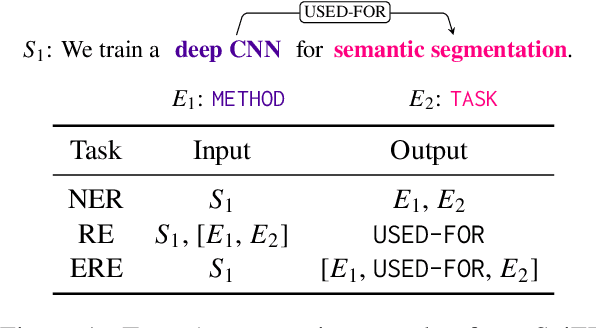
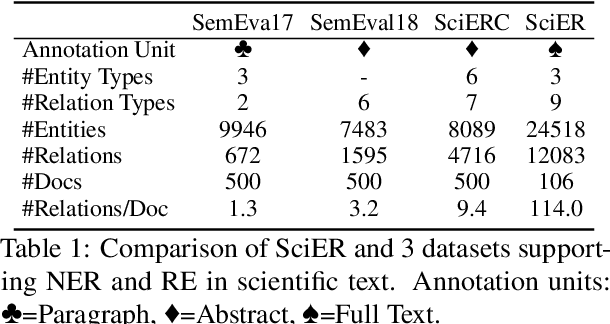


Abstract:Scientific information extraction (SciIE) is critical for converting unstructured knowledge from scholarly articles into structured data (entities and relations). Several datasets have been proposed for training and validating SciIE models. However, due to the high complexity and cost of annotating scientific texts, those datasets restrict their annotations to specific parts of paper, such as abstracts, resulting in the loss of diverse entity mentions and relations in context. In this paper, we release a new entity and relation extraction dataset for entities related to datasets, methods, and tasks in scientific articles. Our dataset contains 106 manually annotated full-text scientific publications with over 24k entities and 12k relations. To capture the intricate use and interactions among entities in full texts, our dataset contains a fine-grained tag set for relations. Additionally, we provide an out-of-distribution test set to offer a more realistic evaluation. We conduct comprehensive experiments, including state-of-the-art supervised models and our proposed LLM-based baselines, and highlight the challenges presented by our dataset, encouraging the development of innovative models to further the field of SciIE.
FlowLearn: Evaluating Large Vision-Language Models on Flowchart Understanding
Jul 09, 2024



Abstract:Flowcharts are graphical tools for representing complex concepts in concise visual representations. This paper introduces the FlowLearn dataset, a resource tailored to enhance the understanding of flowcharts. FlowLearn contains complex scientific flowcharts and simulated flowcharts. The scientific subset contains 3,858 flowcharts sourced from scientific literature and the simulated subset contains 10,000 flowcharts created using a customizable script. The dataset is enriched with annotations for visual components, OCR, Mermaid code representation, and VQA question-answer pairs. Despite the proven capabilities of Large Vision-Language Models (LVLMs) in various visual understanding tasks, their effectiveness in decoding flowcharts - a crucial element of scientific communication - has yet to be thoroughly investigated. The FlowLearn test set is crafted to assess the performance of LVLMs in flowchart comprehension. Our study thoroughly evaluates state-of-the-art LVLMs, identifying existing limitations and establishing a foundation for future enhancements in this relatively underexplored domain. For instance, in tasks involving simulated flowcharts, GPT-4V achieved the highest accuracy (58%) in counting the number of nodes, while Claude recorded the highest accuracy (83%) in OCR tasks. Notably, no single model excels in all tasks within the FlowLearn framework, highlighting significant opportunities for further development.
SciDMT: A Large-Scale Corpus for Detecting Scientific Mentions
Jun 20, 2024



Abstract:We present SciDMT, an enhanced and expanded corpus for scientific mention detection, offering a significant advancement over existing related resources. SciDMT contains annotated scientific documents for datasets (D), methods (M), and tasks (T). The corpus consists of two components: 1) the SciDMT main corpus, which includes 48 thousand scientific articles with over 1.8 million weakly annotated mention annotations in the format of in-text span, and 2) an evaluation set, which comprises 100 scientific articles manually annotated for evaluation purposes. To the best of our knowledge, SciDMT is the largest corpus for scientific entity mention detection. The corpus's scale and diversity are instrumental in developing and refining models for tasks such as indexing scientific papers, enhancing information retrieval, and improving the accessibility of scientific knowledge. We demonstrate the corpus's utility through experiments with advanced deep learning architectures like SciBERT and GPT-3.5. Our findings establish performance baselines and highlight unresolved challenges in scientific mention detection. SciDMT serves as a robust benchmark for the research community, encouraging the development of innovative models to further the field of scientific information extraction.
* LREC/COLING 2024
DMDD: A Large-Scale Dataset for Dataset Mentions Detection
May 19, 2023Abstract:The recognition of dataset names is a critical task for automatic information extraction in scientific literature, enabling researchers to understand and identify research opportunities. However, existing corpora for dataset mention detection are limited in size and naming diversity. In this paper, we introduce the Dataset Mentions Detection Dataset (DMDD), the largest publicly available corpus for this task. DMDD consists of the DMDD main corpus, comprising 31,219 scientific articles with over 449,000 dataset mentions weakly annotated in the format of in-text spans, and an evaluation set, which comprises of 450 scientific articles manually annotated for evaluation purposes. We use DMDD to establish baseline performance for dataset mention detection and linking. By analyzing the performance of various models on DMDD, we are able to identify open problems in dataset mention detection. We invite the community to use our dataset as a challenge to develop novel dataset mention detection models.
Prostate Segmentation from 3D MRI Using a Two-Stage Model and Variable-Input Based Uncertainty Measure
Mar 06, 2019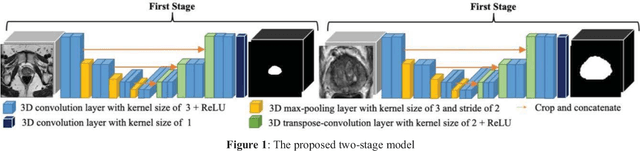
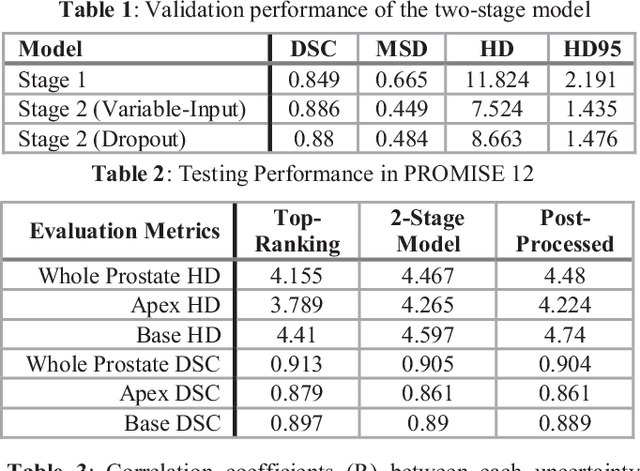
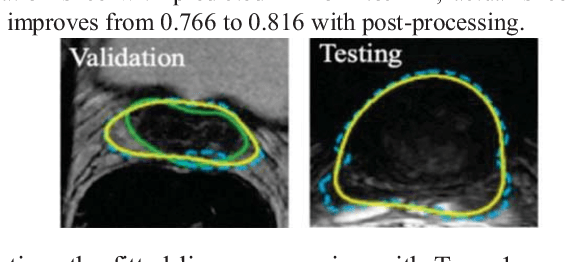
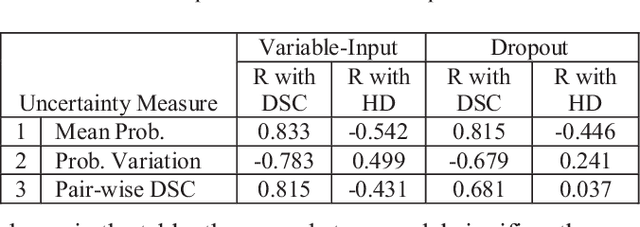
Abstract:This paper proposes a two-stage segmentation model, variable-input based uncertainty measures and an uncertainty-guided post-processing method for prostate segmentation on 3D magnetic resonance images (MRI). The two-stage model was based on 3D dilated U-Nets with the first stage to localize the prostate and the second stage to obtain an accurate segmentation from cropped images. For data augmentation, we proposed the variable-input method which crops the region of interest with additional random variations. Similar to other deep learning models, the proposed model also faced the challenge of suboptimal performance in certain testing cases due to varied training and testing image characteristics. Therefore, it is valuable to evaluate the confidence and performance of the network using uncertainty measures, which are often calculated from the probability maps or their standard deviations with multiple model outputs for the same testing case. However, few studies have quantitatively compared different methods of uncertainty calculation. Furthermore, unlike the commonly used Bayesian dropout during testing, we developed uncertainty measures based on the variable input images at the second stage and evaluated its performance by calculating the correlation with ground-truth-based performance metrics, such as Dice score. For performance estimation, we predicted Dice scores and Hausdorff distance with the most correlated uncertainty measure. For post-processing, we performed Gaussian filter on the underperformed slices to improve segmentation quality. Using PROMISE-12 data, we demonstrated the robustness of the two-stage model and showed high correlation of the proposed variable-input based uncertainty measures with GT-based performance. The uncertainty-guided post-processing method significantly improved label smoothness.
A Self-Adaptive Network For Multiple Sclerosis Lesion Segmentation From Multi-Contrast MRI With Various Imaging Protocols
Nov 19, 2018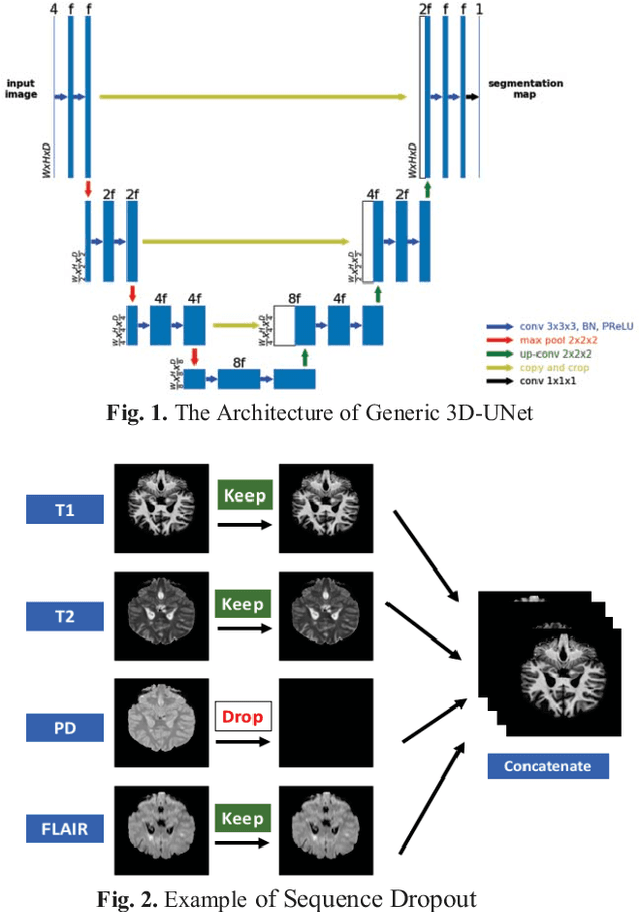
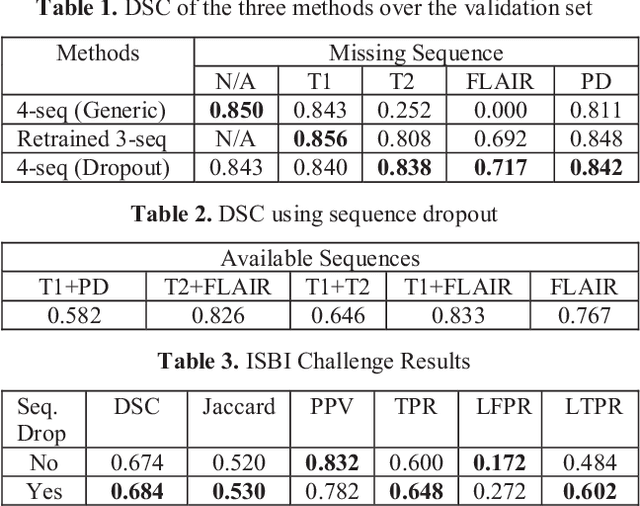
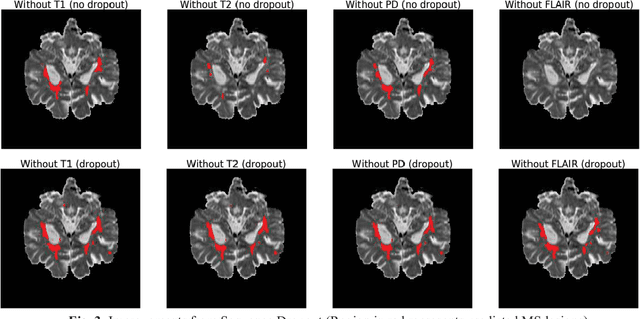
Abstract:Deep neural networks (DNN) have shown promises in the lesion segmentation of multiple sclerosis (MS) from multicontrast MRI including T1, T2, proton density (PD) and FLAIR sequences. However, one challenge in deploying such networks into clinical practice is the variability of imaging protocols, which often differ from the training dataset as certain MRI sequences may be unavailable or unusable. Therefore, trained networks need to adapt to practical situations when imaging protocols are different in deployment. In this paper, we propose a DNN-based MS lesion segmentation framework with a novel technique called sequence dropout which can adapt to various combinations of input MRI sequences during deployment and achieve the maximal possible performance from the given input. In addition, with this framework, we studied the quantitative impact of each MRI sequence on the MS lesion segmentation task without training separate networks. Experiments were performed using the IEEE ISBI 2015 Longitudinal MS Lesion Challenge dataset and our method is currently ranked 2nd with a Dice similarity coefficient of 0.684. Furthermore, we showed our network achieved the maximal possible performance when one sequence is unavailable during deployment by comparing with separate networks trained on the corresponding input MRI sequences. In particular, we discovered T1 and PD have minor impact on segmentation performance while FLAIR is the predominant sequence. Experiments with multiple missing sequences were also performed and showed the robustness of our network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge