A Self-Adaptive Network For Multiple Sclerosis Lesion Segmentation From Multi-Contrast MRI With Various Imaging Protocols
Paper and Code
Nov 19, 2018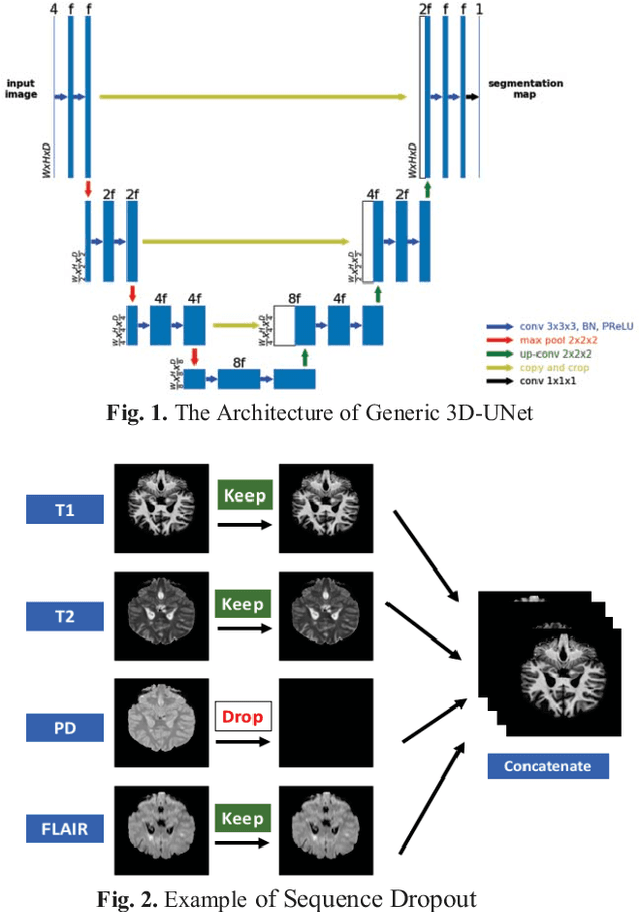
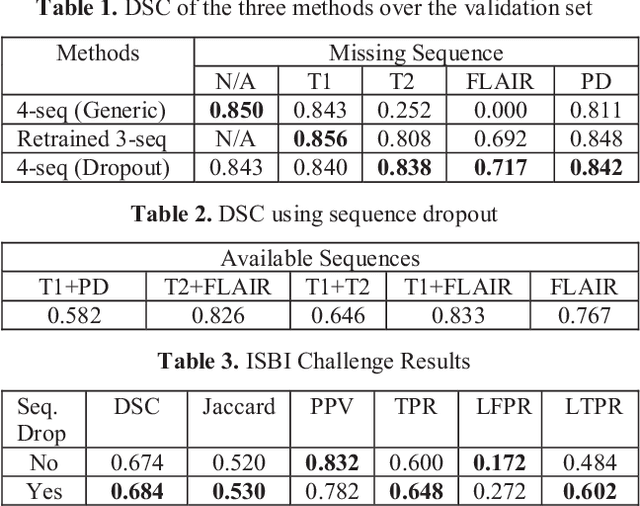
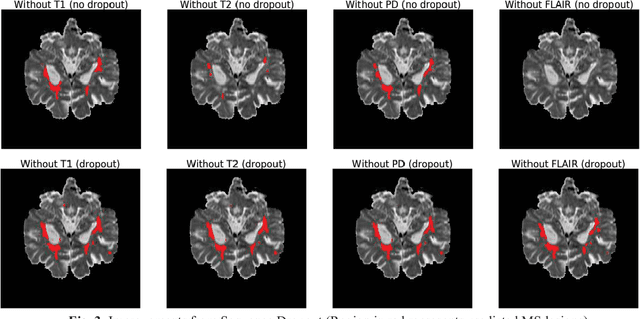
Deep neural networks (DNN) have shown promises in the lesion segmentation of multiple sclerosis (MS) from multicontrast MRI including T1, T2, proton density (PD) and FLAIR sequences. However, one challenge in deploying such networks into clinical practice is the variability of imaging protocols, which often differ from the training dataset as certain MRI sequences may be unavailable or unusable. Therefore, trained networks need to adapt to practical situations when imaging protocols are different in deployment. In this paper, we propose a DNN-based MS lesion segmentation framework with a novel technique called sequence dropout which can adapt to various combinations of input MRI sequences during deployment and achieve the maximal possible performance from the given input. In addition, with this framework, we studied the quantitative impact of each MRI sequence on the MS lesion segmentation task without training separate networks. Experiments were performed using the IEEE ISBI 2015 Longitudinal MS Lesion Challenge dataset and our method is currently ranked 2nd with a Dice similarity coefficient of 0.684. Furthermore, we showed our network achieved the maximal possible performance when one sequence is unavailable during deployment by comparing with separate networks trained on the corresponding input MRI sequences. In particular, we discovered T1 and PD have minor impact on segmentation performance while FLAIR is the predominant sequence. Experiments with multiple missing sequences were also performed and showed the robustness of our network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge