Brain Tumor Segmentation using an Ensemble of 3D U-Nets and Overall Survival Prediction using Radiomic Features
Paper and Code
Dec 03, 2018
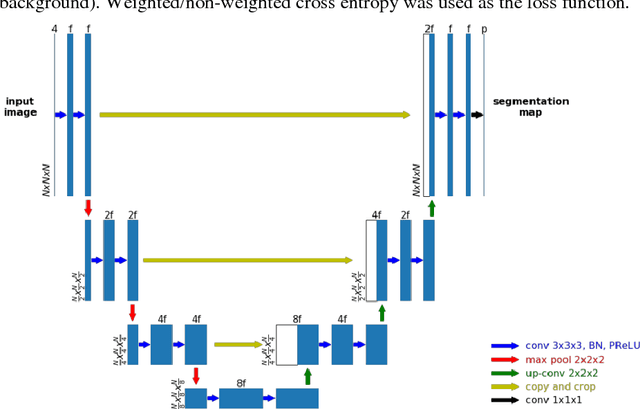
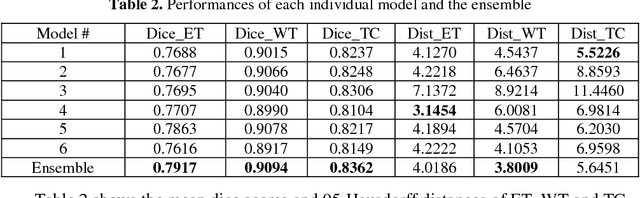
Accurate segmentation of different sub-regions of gliomas including peritumoral edema, necrotic core, enhancing and non-enhancing tumor core from multimodal MRI scans has important clinical relevance in diagnosis, prognosis and treatment of brain tumors. However, due to the highly heterogeneous appearance and shape, segmentation of the sub-regions is very challenging. Recent development using deep learning models has proved its effectiveness in the past several brain segmentation challenges as well as other semantic and medical image segmentation problems. Most models in brain tumor segmentation use a 2D/3D patch to predict the class label for the center voxel and variant patch sizes and scales are used to improve the model performance. However, it has low computation efficiency and also has limited receptive field. U-Net is a widely used network structure for end-to-end segmentation and can be used on the entire image or extracted patches to provide classification labels over the entire input voxels so that it is more efficient and expect to yield better performance with larger input size. Furthermore, instead of picking the best network structure, an ensemble of multiple models, trained on different dataset or different hyper-parameters, can generally improve the segmentation performance. In this study we propose to use an ensemble of 3D U-Nets with different hyper-parameters for brain tumor segmentation. Preliminary results showed effectiveness of this model. In addition, we developed a linear model for survival prediction using extracted imaging and non-imaging features, which, despite the simplicity, can effectively reduce overfitting and regression errors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge