Chee Keong Kwoh
FOCUS: Frequency-Optimized Conditioning of DiffUSion Models for mitigating catastrophic forgetting during Test-Time Adaptation
Aug 20, 2025Abstract:Test-time adaptation enables models to adapt to evolving domains. However, balancing the tradeoff between preserving knowledge and adapting to domain shifts remains challenging for model adaptation methods, since adapting to domain shifts can induce forgetting of task-relevant knowledge. To address this problem, we propose FOCUS, a novel frequency-based conditioning approach within a diffusion-driven input-adaptation framework. Utilising learned, spatially adaptive frequency priors, our approach conditions the reverse steps during diffusion-driven denoising to preserve task-relevant semantic information for dense prediction. FOCUS leverages a trained, lightweight, Y-shaped Frequency Prediction Network (Y-FPN) that disentangles high and low frequency information from noisy images. This minimizes the computational costs involved in implementing our approach in a diffusion-driven framework. We train Y-FPN with FrequencyMix, a novel data augmentation method that perturbs the images across diverse frequency bands, which improves the robustness of our approach to diverse corruptions. We demonstrate the effectiveness of FOCUS for semantic segmentation and monocular depth estimation across 15 corruption types and three datasets, achieving state-of-the-art averaged performance. In addition to improving standalone performance, FOCUS complements existing model adaptation methods since we can derive pseudo labels from FOCUS-denoised images for additional supervision. Even under limited, intermittent supervision with the pseudo labels derived from the FOCUS denoised images, we show that FOCUS mitigates catastrophic forgetting for recent model adaptation methods.
Adapting LLMs to Time Series Forecasting via Temporal Heterogeneity Modeling and Semantic Alignment
Aug 10, 2025Abstract:Large Language Models (LLMs) have recently demonstrated impressive capabilities in natural language processing due to their strong generalization and sequence modeling capabilities. However, their direct application to time series forecasting remains challenging due to two fundamental issues: the inherent heterogeneity of temporal patterns and the modality gap between continuous numerical signals and discrete language representations. In this work, we propose TALON, a unified framework that enhances LLM-based forecasting by modeling temporal heterogeneity and enforcing semantic alignment. Specifically, we design a Heterogeneous Temporal Encoder that partitions multivariate time series into structurally coherent segments, enabling localized expert modeling across diverse temporal patterns. To bridge the modality gap, we introduce a Semantic Alignment Module that aligns temporal features with LLM-compatible representations, enabling effective integration of time series into language-based models while eliminating the need for handcrafted prompts during inference. Extensive experiments on seven real-world benchmarks demonstrate that TALON achieves superior performance across all datasets, with average MSE improvements of up to 11\% over recent state-of-the-art methods. These results underscore the effectiveness of incorporating both pattern-aware and semantic-aware designs when adapting LLMs for time series forecasting. The code is available at: https://github.com/syrGitHub/TALON.
Generating Reliable Pixel-Level Labels for Source Free Domain Adaptation
Jul 03, 2023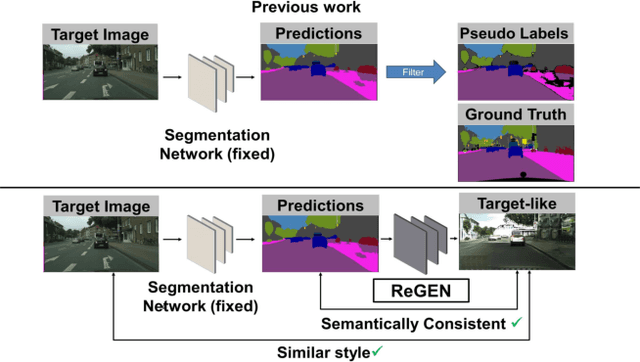
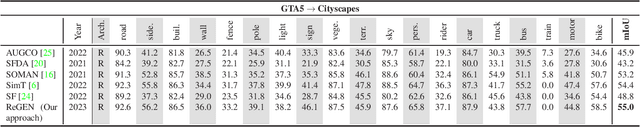
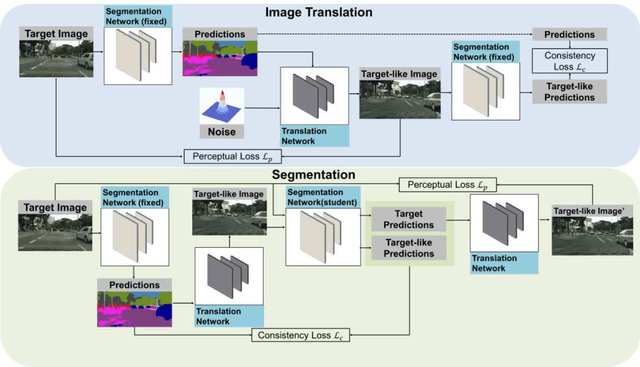
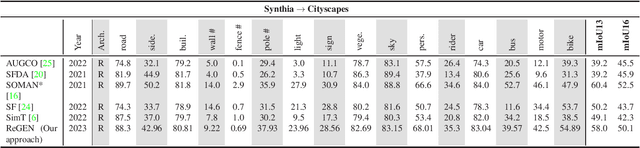
Abstract:This work addresses the challenging domain adaptation setting in which knowledge from the labelled source domain dataset is available only from the pretrained black-box segmentation model. The pretrained model's predictions for the target domain images are noisy because of the distributional differences between the source domain data and the target domain data. Since the model's predictions serve as pseudo labels during self-training, the noise in the predictions impose an upper bound on model performance. Therefore, we propose a simple yet novel image translation workflow, ReGEN, to address this problem. ReGEN comprises an image-to-image translation network and a segmentation network. Our workflow generates target-like images using the noisy predictions from the original target domain images. These target-like images are semantically consistent with the noisy model predictions and therefore can be used to train the segmentation network. In addition to being semantically consistent with the predictions from the original target domain images, the generated target-like images are also stylistically similar to the target domain images. This allows us to leverage the stylistic differences between the target-like images and the target domain image as an additional source of supervision while training the segmentation model. We evaluate our model with two benchmark domain adaptation settings and demonstrate that our approach performs favourably relative to recent state-of-the-art work. The source code will be made available.
Time-Series Representation Learning via Temporal and Contextual Contrasting
Jun 26, 2021



Abstract:Learning decent representations from unlabeled time-series data with temporal dynamics is a very challenging task. In this paper, we propose an unsupervised Time-Series representation learning framework via Temporal and Contextual Contrasting (TS-TCC), to learn time-series representation from unlabeled data. First, the raw time-series data are transformed into two different yet correlated views by using weak and strong augmentations. Second, we propose a novel temporal contrasting module to learn robust temporal representations by designing a tough cross-view prediction task. Last, to further learn discriminative representations, we propose a contextual contrasting module built upon the contexts from the temporal contrasting module. It attempts to maximize the similarity among different contexts of the same sample while minimizing similarity among contexts of different samples. Experiments have been carried out on three real-world time-series datasets. The results manifest that training a linear classifier on top of the features learned by our proposed TS-TCC performs comparably with the supervised training. Additionally, our proposed TS-TCC shows high efficiency in few-labeled data and transfer learning scenarios. The code is publicly available at https://github.com/emadeldeen24/TS-TCC.
COMET: Convolutional Dimension Interaction for Deep Matrix Factorization
Aug 18, 2020



Abstract:Latent factor models play a dominant role among recommendation techniques. However, most of the existing latent factor models assume embedding dimensions are independent of each other, and thus regrettably ignore the interaction information across different embedding dimensions. In this paper, we propose a novel latent factor model called COMET (COnvolutional diMEnsion inTeraction), which provides the first attempt to model higher-order interaction signals among all latent dimensions in an explicit manner. To be specific, COMET stacks the embeddings of historical interactions horizontally, which results in two "embedding maps" that encode the original dimension information. In this way, users' and items' internal interactions can be exploited by convolutional neural networks with kernels of different sizes and a fully-connected multi-layer perceptron. Furthermore, the representations of users and items are enriched by the learnt interaction vectors, which can further be used to produce the final prediction. Extensive experiments and ablation studies on various public implicit feedback datasets clearly demonstrate the effectiveness and the rationality of our proposed method.
Recent Advances in Network-based Methods for Disease Gene Prediction
Jul 19, 2020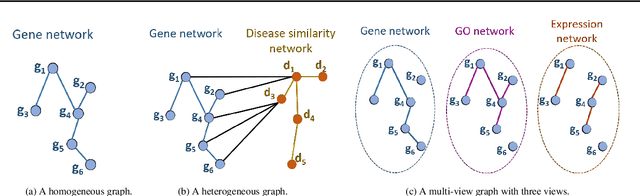
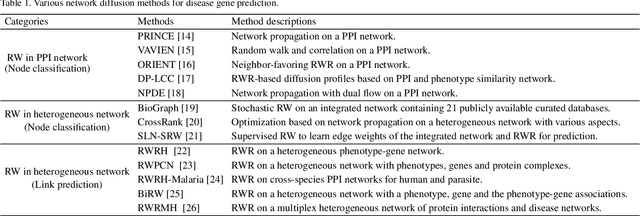
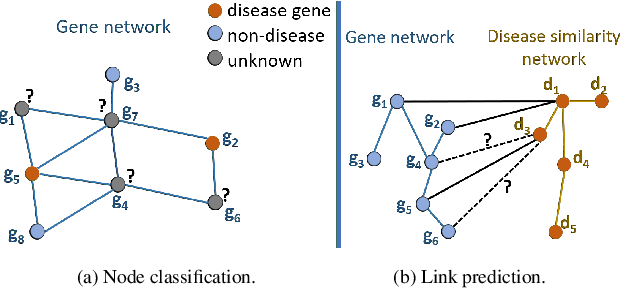
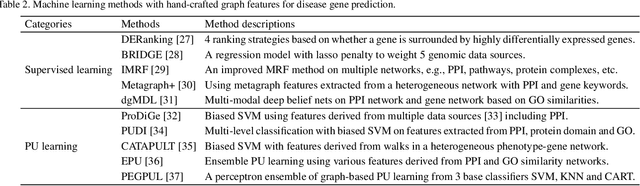
Abstract:Disease-gene association through Genome-wide association study (GWAS) is an arduous task for researchers. Investigating single nucleotide polymorphisms (SNPs) that correlate with specific diseases needs statistical analysis of associations, considering the huge number of possible disease mutations. The most important drawback of GWAS analysis in addition to its high cost is the large number of false-positives. Thus, researchers search for more evidence to cross-check their results through different sources. To provide the researchers with alternative low-cost disease-gene association evidence, computational approaches come into play. Since molecular networks are able to capture complex interplay among molecules in diseases, they become one of the most extensively used data for disease-gene association prediction. In this survey, we aim to provide a comprehensive and an up-to-date review of network-based methods for disease gene prediction. We also conduct an empirical analysis on 14 state-of-the-art methods. To summarize, we first elucidate the task definition for disease gene prediction. Secondly, we categorize existing network-based efforts into network diffusion methods, traditional machine learning methods with handcrafted graph features and graph representation learning methods. Thirdly, an empirical analysis is conducted to evaluate the performance of the selected methods across seven diseases. We also provide distinguishing findings about the discussed methods based on our empirical analysis. Finally, we highlight potential research directions for future studies on disease gene prediction.
Multi-View Collaborative Network Embedding
May 17, 2020


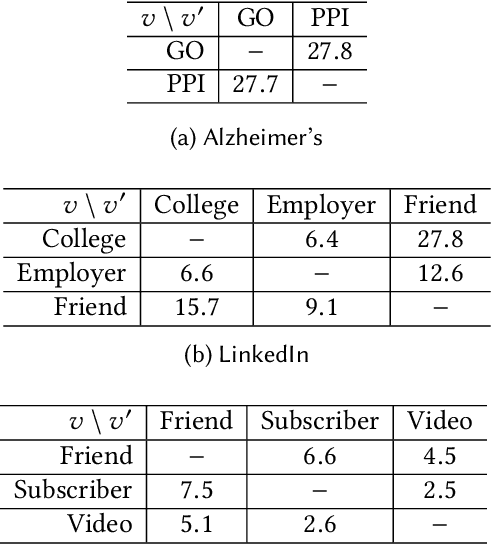
Abstract:Real-world networks often exist with multiple views, where each view describes one type of interaction among a common set of nodes. For example, on a video-sharing network, while two user nodes are linked if they have common favorite videos in one view, they can also be linked in another view if they share common subscribers. Unlike traditional single-view networks, multiple views maintain different semantics to complement each other. In this paper, we propose MANE, a multi-view network embedding approach to learn low-dimensional representations. Similar to existing studies, MANE hinges on diversity and collaboration - while diversity enables views to maintain their individual semantics, collaboration enables views to work together. However, we also discover a novel form of second-order collaboration that has not been explored previously, and further unify it into our framework to attain superior node representations. Furthermore, as each view often has varying importance w.r.t. different nodes, we propose MANE+, an attention-based extension of MANE to model node-wise view importance. Finally, we conduct comprehensive experiments on three public, real-world multi-view networks, and the results demonstrate that our models consistently outperform state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge