Sezin Kircali Ata
Recent Advances in Network-based Methods for Disease Gene Prediction
Jul 19, 2020
Abstract:Disease-gene association through Genome-wide association study (GWAS) is an arduous task for researchers. Investigating single nucleotide polymorphisms (SNPs) that correlate with specific diseases needs statistical analysis of associations, considering the huge number of possible disease mutations. The most important drawback of GWAS analysis in addition to its high cost is the large number of false-positives. Thus, researchers search for more evidence to cross-check their results through different sources. To provide the researchers with alternative low-cost disease-gene association evidence, computational approaches come into play. Since molecular networks are able to capture complex interplay among molecules in diseases, they become one of the most extensively used data for disease-gene association prediction. In this survey, we aim to provide a comprehensive and an up-to-date review of network-based methods for disease gene prediction. We also conduct an empirical analysis on 14 state-of-the-art methods. To summarize, we first elucidate the task definition for disease gene prediction. Secondly, we categorize existing network-based efforts into network diffusion methods, traditional machine learning methods with handcrafted graph features and graph representation learning methods. Thirdly, an empirical analysis is conducted to evaluate the performance of the selected methods across seven diseases. We also provide distinguishing findings about the discussed methods based on our empirical analysis. Finally, we highlight potential research directions for future studies on disease gene prediction.
Multi-View Collaborative Network Embedding
May 17, 2020


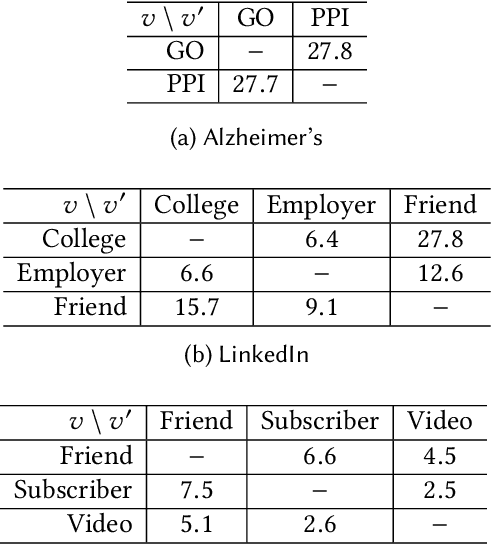
Abstract:Real-world networks often exist with multiple views, where each view describes one type of interaction among a common set of nodes. For example, on a video-sharing network, while two user nodes are linked if they have common favorite videos in one view, they can also be linked in another view if they share common subscribers. Unlike traditional single-view networks, multiple views maintain different semantics to complement each other. In this paper, we propose MANE, a multi-view network embedding approach to learn low-dimensional representations. Similar to existing studies, MANE hinges on diversity and collaboration - while diversity enables views to maintain their individual semantics, collaboration enables views to work together. However, we also discover a novel form of second-order collaboration that has not been explored previously, and further unify it into our framework to attain superior node representations. Furthermore, as each view often has varying importance w.r.t. different nodes, we propose MANE+, an attention-based extension of MANE to model node-wise view importance. Finally, we conduct comprehensive experiments on three public, real-world multi-view networks, and the results demonstrate that our models consistently outperform state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge