Cameron Diao
Shammie
CodonMPNN for Organism Specific and Codon Optimal Inverse Folding
Sep 25, 2024Abstract:Generating protein sequences conditioned on protein structures is an impactful technique for protein engineering. When synthesizing engineered proteins, they are commonly translated into DNA and expressed in an organism such as yeast. One difficulty in this process is that the expression rates can be low due to suboptimal codon sequences for expressing a protein in a host organism. We propose CodonMPNN, which generates a codon sequence conditioned on a protein backbone structure and an organism label. If naturally occurring DNA sequences are close to codon optimality, CodonMPNN could learn to generate codon sequences with higher expression yields than heuristic codon choices for generated amino acid sequences. Experiments show that CodonMPNN retains the performance of previous inverse folding approaches and recovers wild-type codons more frequently than baselines. Furthermore, CodonMPNN has a higher likelihood of generating high-fitness codon sequences than low-fitness codon sequences for the same protein sequence. Code is available at https://github.com/HannesStark/CodonMPNN.
MolCPT: Molecule Continuous Prompt Tuning to Generalize Molecular Representation Learning
Dec 20, 2022Abstract:Molecular representation learning is crucial for the problem of molecular property prediction, where graph neural networks (GNNs) serve as an effective solution due to their structure modeling capabilities. Since labeled data is often scarce and expensive to obtain, it is a great challenge for GNNs to generalize in the extensive molecular space. Recently, the training paradigm of "pre-train, fine-tune" has been leveraged to improve the generalization capabilities of GNNs. It uses self-supervised information to pre-train the GNN, and then performs fine-tuning to optimize the downstream task with just a few labels. However, pre-training does not always yield statistically significant improvement, especially for self-supervised learning with random structural masking. In fact, the molecular structure is characterized by motif subgraphs, which are frequently occurring and influence molecular properties. To leverage the task-related motifs, we propose a novel paradigm of "pre-train, prompt, fine-tune" for molecular representation learning, named molecule continuous prompt tuning (MolCPT). MolCPT defines a motif prompting function that uses the pre-trained model to project the standalone input into an expressive prompt. The prompt effectively augments the molecular graph with meaningful motifs in the continuous representation space; this provides more structural patterns to aid the downstream classifier in identifying molecular properties. Extensive experiments on several benchmark datasets show that MolCPT efficiently generalizes pre-trained GNNs for molecular property prediction, with or without a few fine-tuning steps.
Relational Attention: Generalizing Transformers for Graph-Structured Tasks
Oct 11, 2022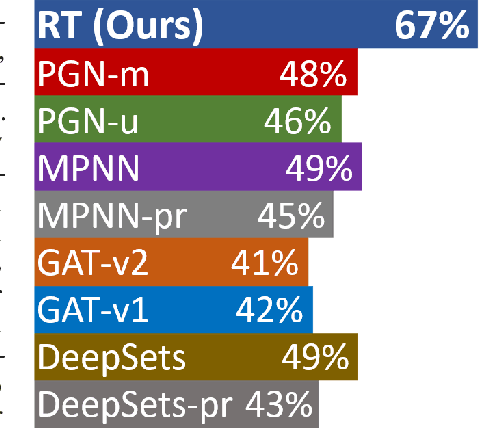
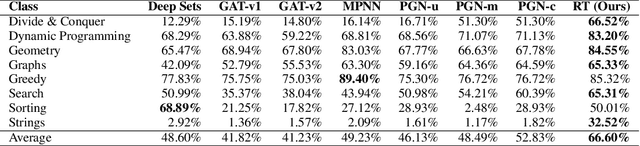
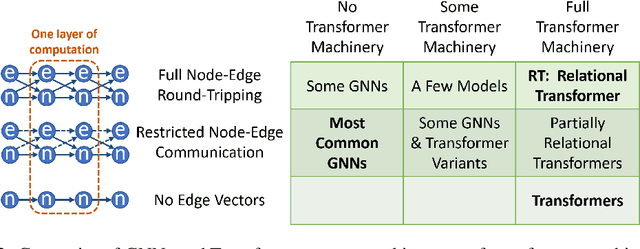
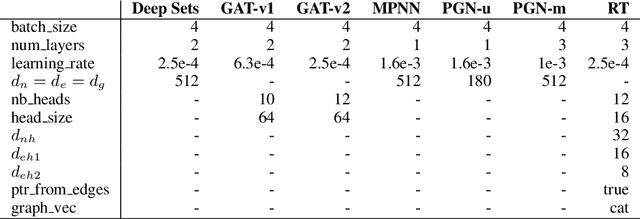
Abstract:Transformers flexibly operate over sets of real-valued vectors representing task-specific entities and their attributes, where each vector might encode one word-piece token and its position in a sequence, or some piece of information that carries no position at all. But as set processors, transformers are at a disadvantage in reasoning over more general graph-structured data where nodes represent entities and edges represent relations between entities. To address this shortcoming, we generalize transformer attention to consider and update edge vectors in each transformer layer. We evaluate this relational transformer on a diverse array of graph-structured tasks, including the large and challenging CLRS Algorithmic Reasoning Benchmark. There, it dramatically outperforms state-of-the-art graph neural networks expressly designed to reason over graph-structured data. Our analysis demonstrates that these gains are attributable to relational attention's inherent ability to leverage the greater expressivity of graphs over sets.
Beyond the Imitation Game: Quantifying and extrapolating the capabilities of language models
Jun 10, 2022Abstract:Language models demonstrate both quantitative improvement and new qualitative capabilities with increasing scale. Despite their potentially transformative impact, these new capabilities are as yet poorly characterized. In order to inform future research, prepare for disruptive new model capabilities, and ameliorate socially harmful effects, it is vital that we understand the present and near-future capabilities and limitations of language models. To address this challenge, we introduce the Beyond the Imitation Game benchmark (BIG-bench). BIG-bench currently consists of 204 tasks, contributed by 442 authors across 132 institutions. Task topics are diverse, drawing problems from linguistics, childhood development, math, common-sense reasoning, biology, physics, social bias, software development, and beyond. BIG-bench focuses on tasks that are believed to be beyond the capabilities of current language models. We evaluate the behavior of OpenAI's GPT models, Google-internal dense transformer architectures, and Switch-style sparse transformers on BIG-bench, across model sizes spanning millions to hundreds of billions of parameters. In addition, a team of human expert raters performed all tasks in order to provide a strong baseline. Findings include: model performance and calibration both improve with scale, but are poor in absolute terms (and when compared with rater performance); performance is remarkably similar across model classes, though with benefits from sparsity; tasks that improve gradually and predictably commonly involve a large knowledge or memorization component, whereas tasks that exhibit "breakthrough" behavior at a critical scale often involve multiple steps or components, or brittle metrics; social bias typically increases with scale in settings with ambiguous context, but this can be improved with prompting.
Generalized Learning Vector Quantization for Classification in Randomized Neural Networks and Hyperdimensional Computing
Jun 17, 2021



Abstract:Machine learning algorithms deployed on edge devices must meet certain resource constraints and efficiency requirements. Random Vector Functional Link (RVFL) networks are favored for such applications due to their simple design and training efficiency. We propose a modified RVFL network that avoids computationally expensive matrix operations during training, thus expanding the network's range of potential applications. Our modification replaces the least-squares classifier with the Generalized Learning Vector Quantization (GLVQ) classifier, which only employs simple vector and distance calculations. The GLVQ classifier can also be considered an improvement upon certain classification algorithms popularly used in the area of Hyperdimensional Computing. The proposed approach achieved state-of-the-art accuracy on a collection of datasets from the UCI Machine Learning Repository - higher than previously proposed RVFL networks. We further demonstrate that our approach still achieves high accuracy while severely limited in training iterations (using on average only 21% of the least-squares classifier computational costs).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge