Audrey G. Chung
Enhancing Food Intake Tracking in Long-Term Care with Automated Food Imaging and Nutrient Intake Tracking Technology
Dec 08, 2021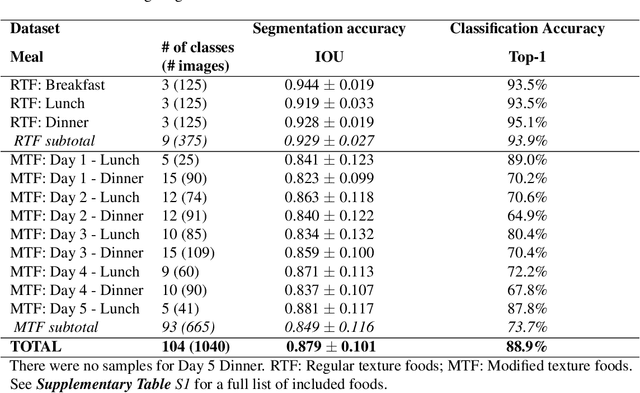
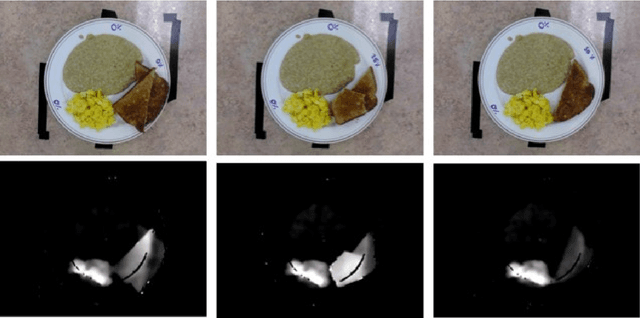
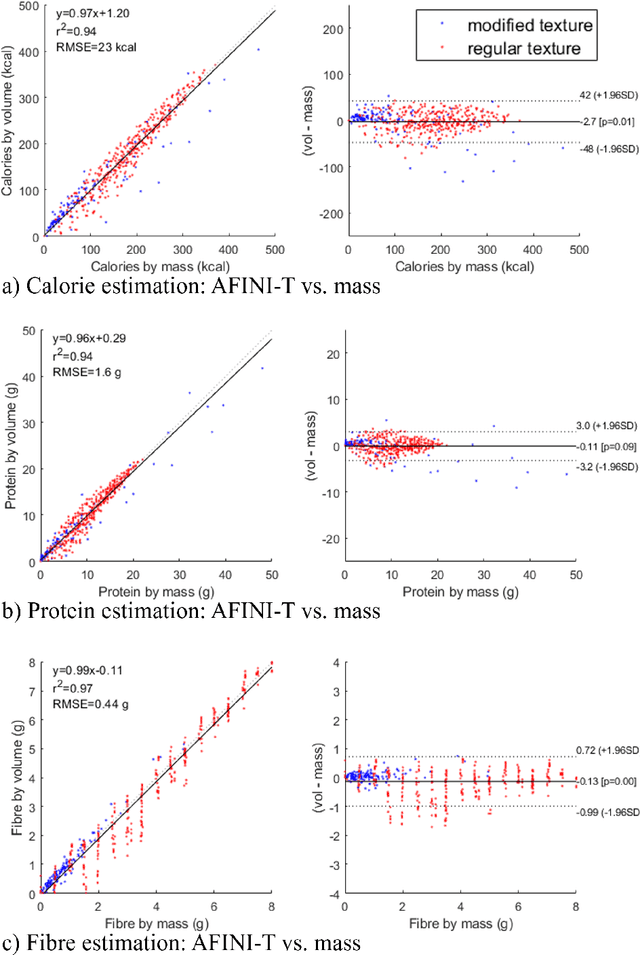
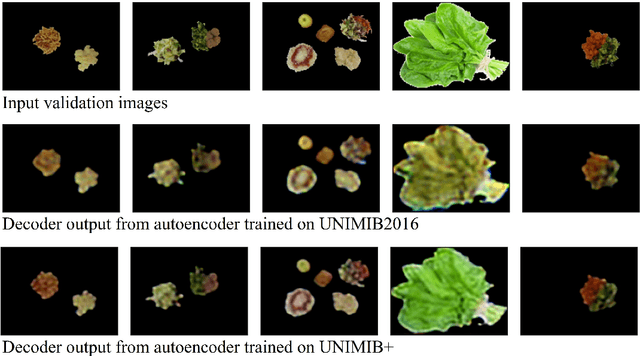
Abstract:Half of long-term care (LTC) residents are malnourished increasing hospitalization, mortality, morbidity, with lower quality of life. Current tracking methods are subjective and time consuming. This paper presents the automated food imaging and nutrient intake tracking (AFINI-T) technology designed for LTC. We propose a novel convolutional autoencoder for food classification, trained on an augmented UNIMIB2016 dataset and tested on our simulated LTC food intake dataset (12 meal scenarios; up to 15 classes each; top-1 classification accuracy: 88.9%; mean intake error: -0.4 mL$\pm$36.7 mL). Nutrient intake estimation by volume was strongly linearly correlated with nutrient estimates from mass ($r^2$ 0.92 to 0.99) with good agreement between methods ($\sigma$= -2.7 to -0.01; zero within each of the limits of agreement). The AFINI-T approach is a deep-learning powered computational nutrient sensing system that may provide a novel means for more accurately and objectively tracking LTC resident food intake to support and prevent malnutrition tracking strategies.
COVID-Net MLSys: Designing COVID-Net for the Clinical Workflow
Sep 14, 2021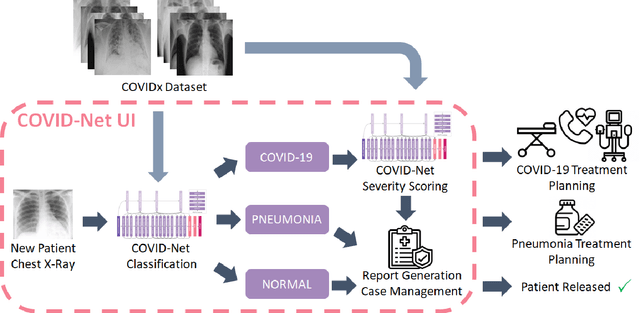
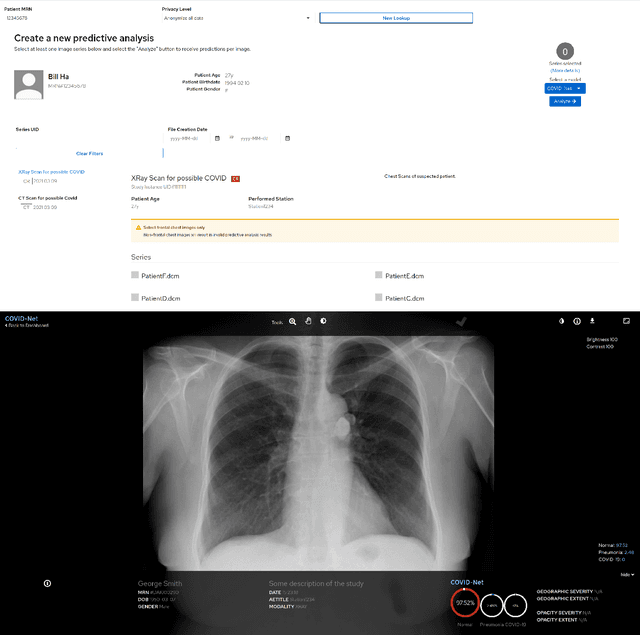
Abstract:As the COVID-19 pandemic continues to devastate globally, one promising field of research is machine learning-driven computer vision to streamline various parts of the COVID-19 clinical workflow. These machine learning methods are typically stand-alone models designed without consideration for the integration necessary for real-world application workflows. In this study, we take a machine learning and systems (MLSys) perspective to design a system for COVID-19 patient screening with the clinical workflow in mind. The COVID-Net system is comprised of the continuously evolving COVIDx dataset, COVID-Net deep neural network for COVID-19 patient detection, and COVID-Net S deep neural networks for disease severity scoring for COVID-19 positive patient cases. The deep neural networks within the COVID-Net system possess state-of-the-art performance, and are designed to be integrated within a user interface (UI) for clinical decision support with automatic report generation to assist clinicians in their treatment decisions.
COVID-Net CXR-2: An Enhanced Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images
May 14, 2021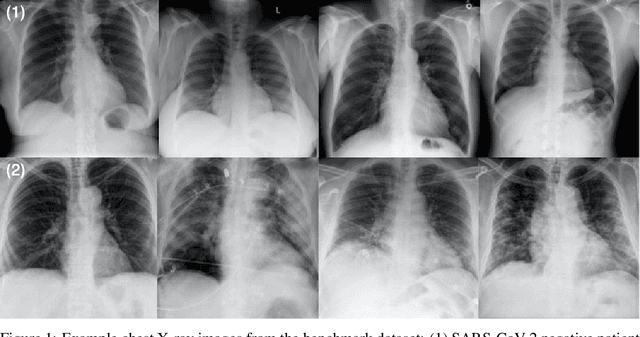
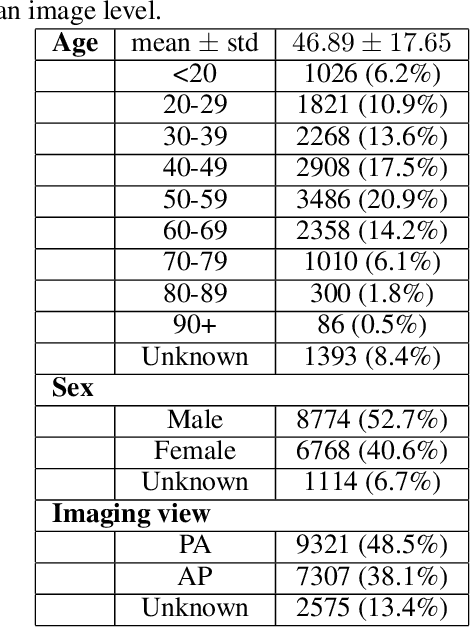
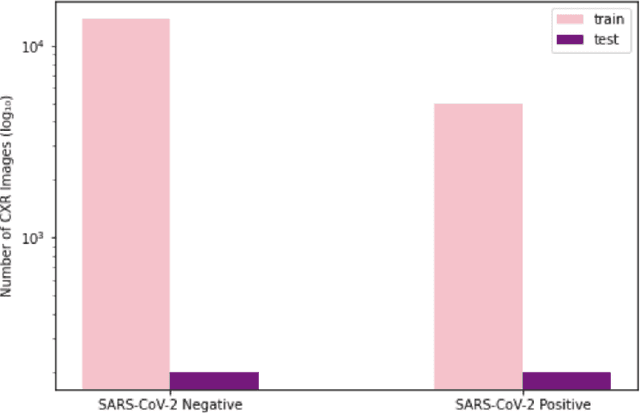
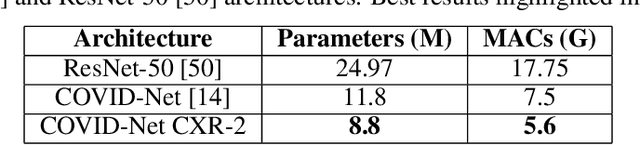
Abstract:As the COVID-19 pandemic continues to devastate globally, the use of chest X-ray (CXR) imaging as a complimentary screening strategy to RT-PCR testing continues to grow given its routine clinical use for respiratory complaint. As part of the COVID-Net open source initiative, we introduce COVID-Net CXR-2, an enhanced deep convolutional neural network design for COVID-19 detection from CXR images built using a greater quantity and diversity of patients than the original COVID-Net. To facilitate this, we also introduce a new benchmark dataset composed of 19,203 CXR images from a multinational cohort of 16,656 patients from at least 51 countries, making it the largest, most diverse COVID-19 CXR dataset in open access form. The COVID-Net CXR-2 network achieves sensitivity and positive predictive value of 95.5%/97.0%, respectively, and was audited in a transparent and responsible manner. Explainability-driven performance validation was used during auditing to gain deeper insights in its decision-making behaviour and to ensure clinically relevant factors are leveraged for improving trust in its usage. Radiologist validation was also conducted, where select cases were reviewed and reported on by two board-certified radiologists with over 10 and 19 years of experience, respectively, and showed that the critical factors leveraged by COVID-Net CXR-2 are consistent with radiologist interpretations. While not a production-ready solution, we hope the open-source, open-access release of COVID-Net CXR-2 and the respective CXR benchmark dataset will encourage researchers, clinical scientists, and citizen scientists to accelerate advancements and innovations in the fight against the pandemic.
Towards computer-aided severity assessment: training and validation of deep neural networks for geographic extent and opacity extent scoring of chest X-rays for SARS-CoV-2 lung disease severity
May 27, 2020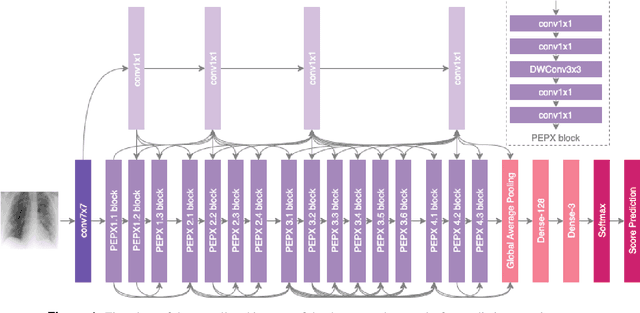
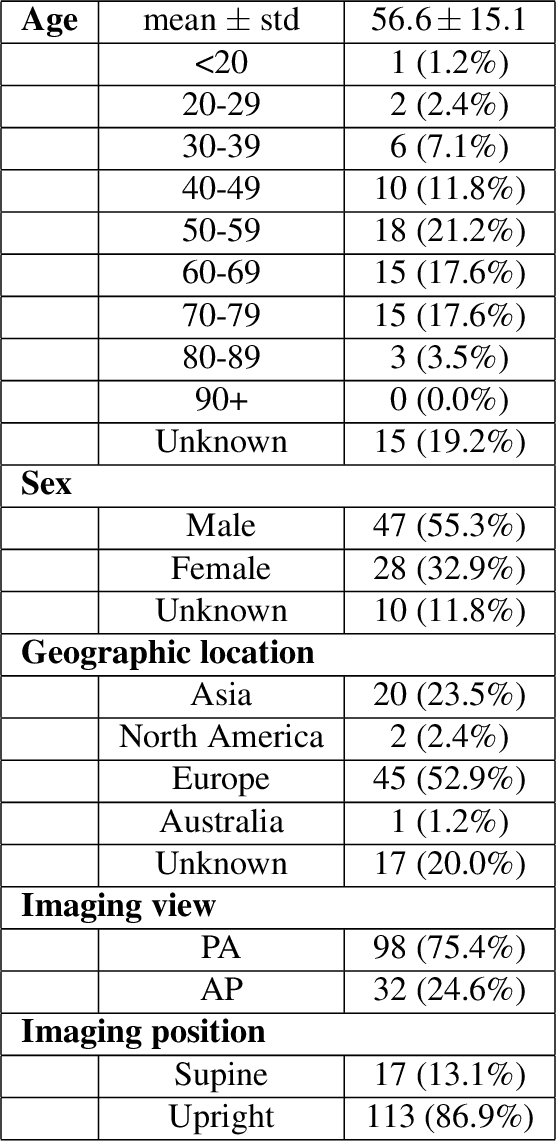
Abstract:Background: A critical step in effective care and treatment planning for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the assessment of the severity of disease progression. Chest x-rays (CXRs) are often used to assess SARS-CoV-2 severity, with two important assessment metrics being extent of lung involvement and degree of opacity. In this proof-of-concept study, we assess the feasibility of computer-aided scoring of CXRs of SARS-CoV-2 lung disease severity using a deep learning system. Materials and Methods: Data consisted of 130 CXRs from SARS-CoV-2 positive patient cases from the Cohen study. Geographic extent and opacity extent were scored by two board-certified expert chest radiologists (with 20+ years of experience) and a 2nd-year radiology resident. The deep neural networks used in this study are based on a COVID-Net network architecture. 100 versions of the network were independently learned (50 to perform geographic extent scoring and 50 to perform opacity extent scoring) using random subsets of CXRs from the Cohen study, and evaluated the networks using stratified Monte Carlo cross-validation experiments. Findings: The deep neural networks yielded R$^2$ of 0.673 $\pm$ 0.004 and 0.636 $\pm$ 0.002 between predicted scores and radiologist scores for geographic extent and opacity extent, respectively, in stratified Monte Carlo cross-validation experiments. The best performing networks achieved R$^2$ of 0.865 and 0.746 between predicted scores and radiologist scores for geographic extent and opacity extent, respectively. Interpretation: The results are promising and suggest that the use of deep neural networks on CXRs could be an effective tool for computer-aided assessment of SARS-CoV-2 lung disease severity, although additional studies are needed before adoption for routine clinical use.
ProstateGAN: Mitigating Data Bias via Prostate Diffusion Imaging Synthesis with Generative Adversarial Networks
Nov 21, 2018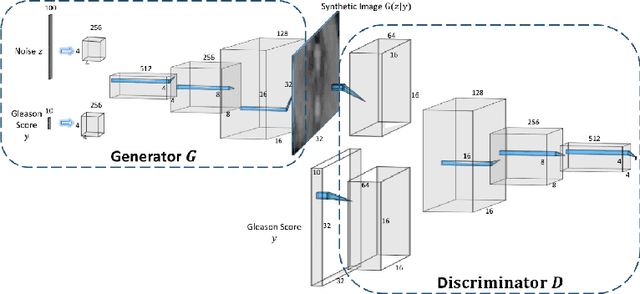
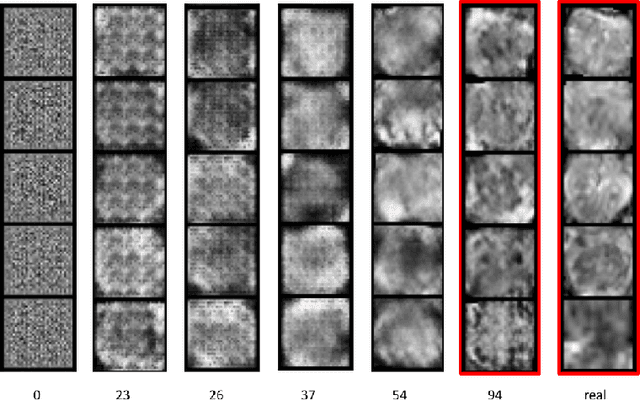
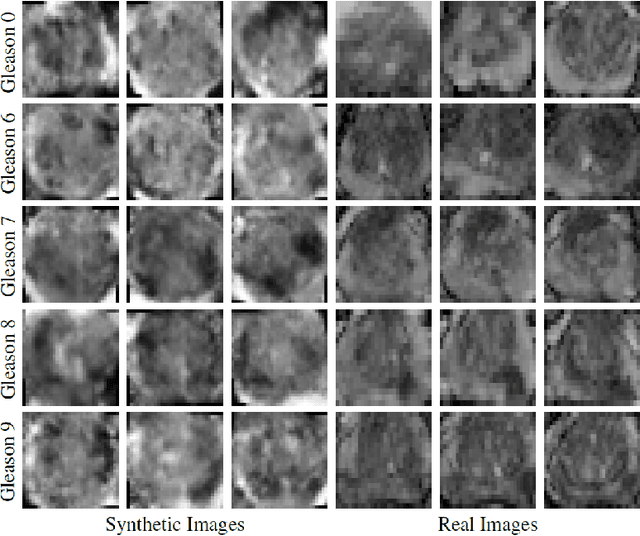
Abstract:Generative Adversarial Networks (GANs) have shown considerable promise for mitigating the challenge of data scarcity when building machine learning-driven analysis algorithms. Specifically, a number of studies have shown that GAN-based image synthesis for data augmentation can aid in improving classification accuracy in a number of medical image analysis tasks, such as brain and liver image analysis. However, the efficacy of leveraging GANs for tackling prostate cancer analysis has not been previously explored. Motivated by this, in this study we introduce ProstateGAN, a GAN-based model for synthesizing realistic prostate diffusion imaging data. More specifically, in order to generate new diffusion imaging data corresponding to a particular cancer grade (Gleason score), we propose a conditional deep convolutional GAN architecture that takes Gleason scores into consideration during the training process. Experimental results show that high-quality synthetic prostate diffusion imaging data can be generated using the proposed ProstateGAN for specified Gleason scores.
EdgeSpeechNets: Highly Efficient Deep Neural Networks for Speech Recognition on the Edge
Oct 18, 2018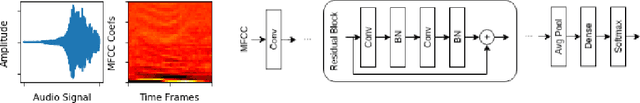
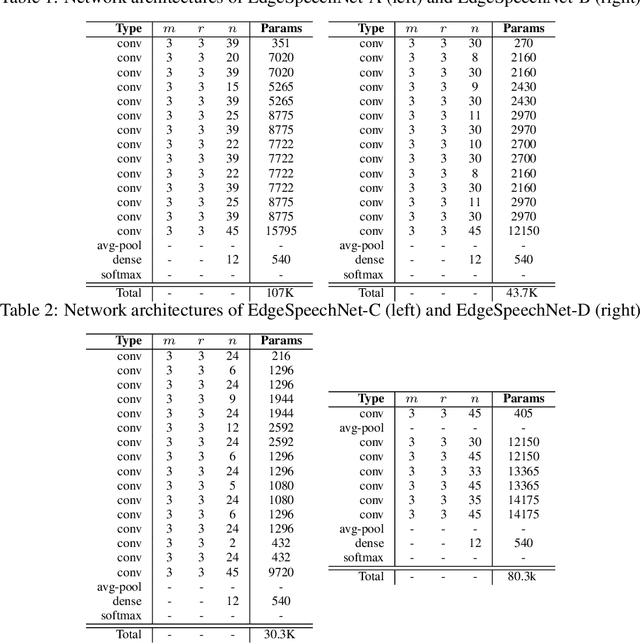
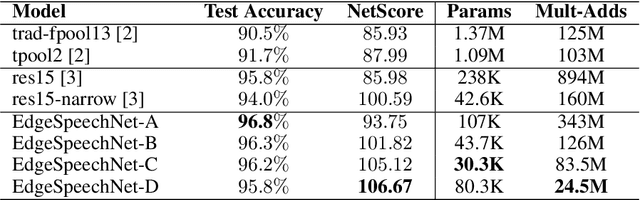
Abstract:Despite showing state-of-the-art performance, deep learning for speech recognition remains challenging to deploy in on-device edge scenarios such as mobile and other consumer devices. Recently, there have been greater efforts in the design of small, low-footprint deep neural networks (DNNs) that are more appropriate for edge devices, with much of the focus on design principles for hand-crafting efficient network architectures. In this study, we explore a human-machine collaborative design strategy for building low-footprint DNN architectures for speech recognition through a marriage of human-driven principled network design prototyping and machine-driven design exploration. The efficacy of this design strategy is demonstrated through the design of a family of highly-efficient DNNs (nicknamed EdgeSpeechNets) for limited-vocabulary speech recognition. Experimental results using the Google Speech Commands dataset for limited-vocabulary speech recognition showed that EdgeSpeechNets have higher accuracies than state-of-the-art DNNs (with the best EdgeSpeechNet achieving ~97% accuracy), while achieving significantly smaller network sizes (as much as 7.8x smaller) and lower computational cost (as much as 36x fewer multiply-add operations, 10x lower prediction latency, and 16x smaller memory footprint on a Motorola Moto E phone), making them very well-suited for on-device edge voice interface applications.
Nature vs. Nurture: The Role of Environmental Resources in Evolutionary Deep Intelligence
Feb 09, 2018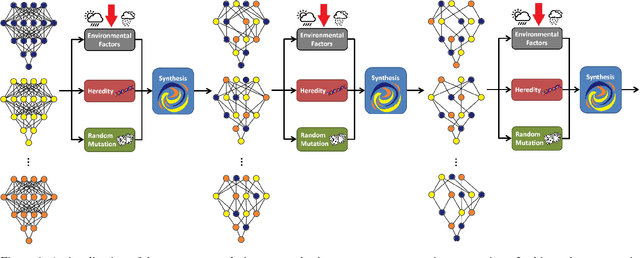

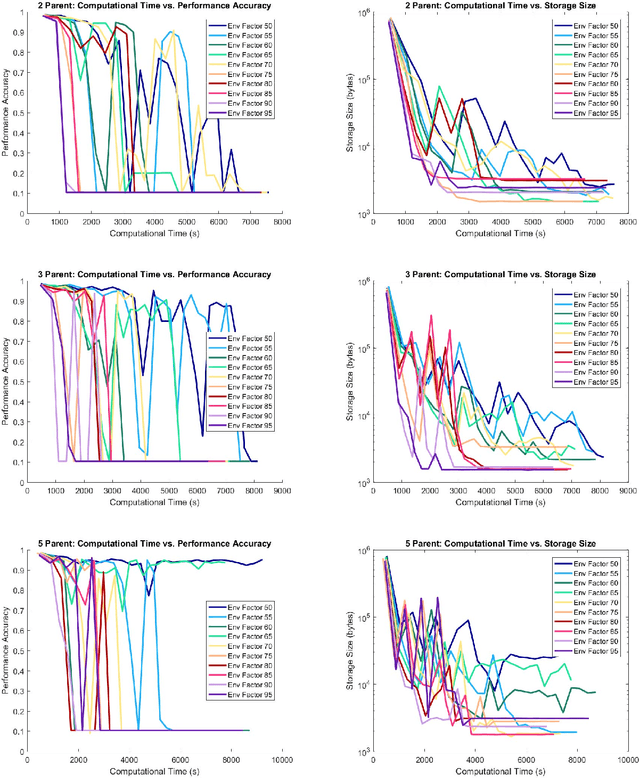
Abstract:Evolutionary deep intelligence synthesizes highly efficient deep neural networks architectures over successive generations. Inspired by the nature versus nurture debate, we propose a study to examine the role of external factors on the network synthesis process by varying the availability of simulated environmental resources. Experimental results were obtained for networks synthesized via asexual evolutionary synthesis (1-parent) and sexual evolutionary synthesis (2-parent, 3-parent, and 5-parent) using a 10% subset of the MNIST dataset. Results show that a lower environmental factor model resulted in a more gradual loss in performance accuracy and decrease in storage size. This potentially allows significantly reduced storage size with minimal to no drop in performance accuracy, and the best networks were synthesized using the lowest environmental factor models.
A new take on measuring relative nutritional density: The feasibility of using a deep neural network to assess commercially-prepared pureed food concentrations
Nov 03, 2017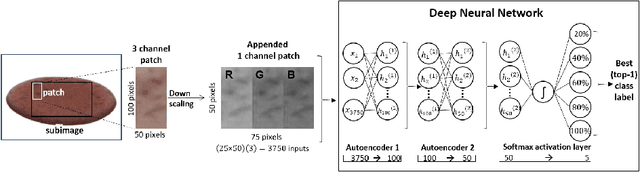
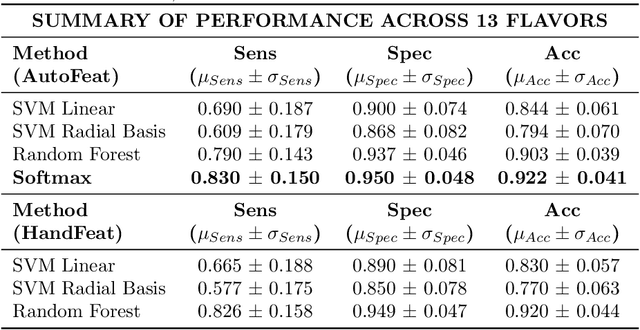
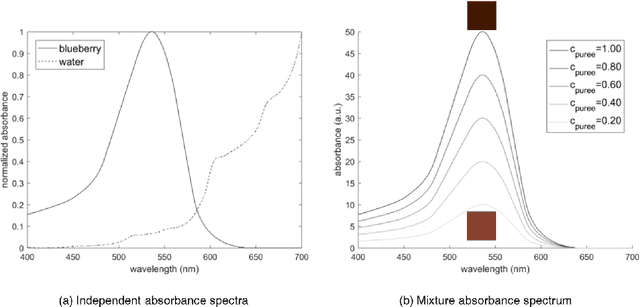
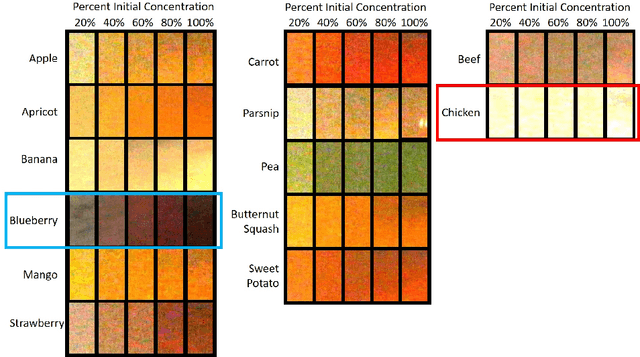
Abstract:Dysphagia affects 590 million people worldwide and increases risk for malnutrition. Pureed food may reduce choking, however preparation differences impact nutrient density making quality assurance necessary. This paper is the first study to investigate the feasibility of computational pureed food nutritional density analysis using an imaging system. Motivated by a theoretical optical dilution model, a novel deep neural network (DNN) was evaluated using 390 samples from thirteen types of commercially prepared purees at five dilutions. The DNN predicted relative concentration of the puree sample (20%, 40%, 60%, 80%, 100% initial concentration). Data were captured using same-side reflectance of multispectral imaging data at different polarizations at three exposures. Experimental results yielded an average top-1 prediction accuracy of 92.2+/-0.41% with sensitivity and specificity of 83.0+/-15.0% and 95.0+/-4.8%, respectively. This DNN imaging system for nutrient density analysis of pureed food shows promise as a novel tool for nutrient quality assurance.
Discovery Radiomics via Evolutionary Deep Radiomic Sequencer Discovery for Pathologically-Proven Lung Cancer Detection
Oct 20, 2017Abstract:While lung cancer is the second most diagnosed form of cancer in men and women, a sufficiently early diagnosis can be pivotal in patient survival rates. Imaging-based, or radiomics-driven, detection methods have been developed to aid diagnosticians, but largely rely on hand-crafted features which may not fully encapsulate the differences between cancerous and healthy tissue. Recently, the concept of discovery radiomics was introduced, where custom abstract features are discovered from readily available imaging data. We propose a novel evolutionary deep radiomic sequencer discovery approach based on evolutionary deep intelligence. Motivated by patient privacy concerns and the idea of operational artificial intelligence, the evolutionary deep radiomic sequencer discovery approach organically evolves increasingly more efficient deep radiomic sequencers that produce significantly more compact yet similarly descriptive radiomic sequences over multiple generations. As a result, this framework improves operational efficiency and enables diagnosis to be run locally at the radiologist's computer while maintaining detection accuracy. We evaluated the evolved deep radiomic sequencer (EDRS) discovered via the proposed evolutionary deep radiomic sequencer discovery framework against state-of-the-art radiomics-driven and discovery radiomics methods using clinical lung CT data with pathologically-proven diagnostic data from the LIDC-IDRI dataset. The evolved deep radiomic sequencer shows improved sensitivity (93.42%), specificity (82.39%), and diagnostic accuracy (88.78%) relative to previous radiomics approaches.
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction
Mar 28, 2017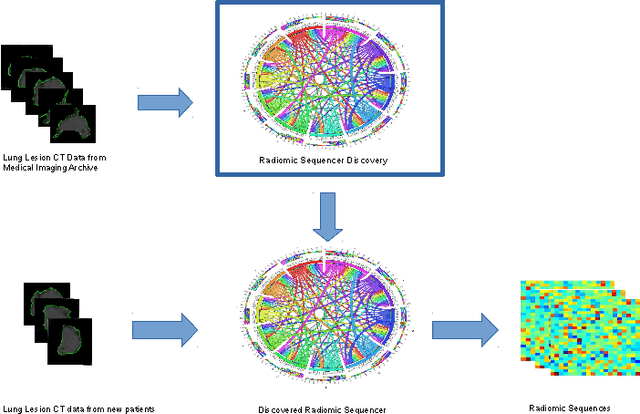
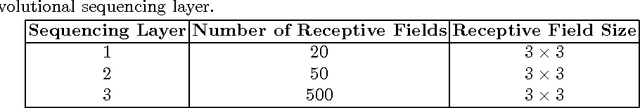
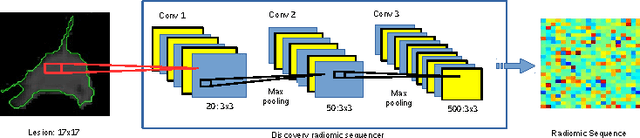
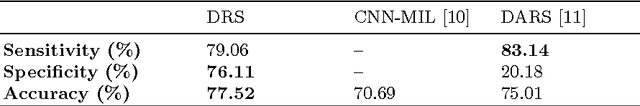
Abstract:Lung cancer is the leading cause for cancer related deaths. As such, there is an urgent need for a streamlined process that can allow radiologists to provide diagnosis with greater efficiency and accuracy. A powerful tool to do this is radiomics: a high-dimension imaging feature set. In this study, we take the idea of radiomics one step further by introducing the concept of discovery radiomics for lung cancer prediction using CT imaging data. In this study, we realize these custom radiomic sequencers as deep convolutional sequencers using a deep convolutional neural network learning architecture. To illustrate the prognostic power and effectiveness of the radiomic sequences produced by the discovered sequencer, we perform cancer prediction between malignant and benign lesions from 97 patients using the pathologically-proven diagnostic data from the LIDC-IDRI dataset. Using the clinically provided pathologically-proven data as ground truth, the proposed framework provided an average accuracy of 77.52% via 10-fold cross-validation with a sensitivity of 79.06% and specificity of 76.11%, surpassing the state-of-the art method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge