Masoom A. Haider
Mitigating Multi-Sequence 3D Prostate MRI Data Scarcity through Domain Adaptation using Locally-Trained Latent Diffusion Models for Prostate Cancer Detection
Jul 08, 2025Abstract:Objective: Latent diffusion models (LDMs) could mitigate data scarcity challenges affecting machine learning development for medical image interpretation. The recent CCELLA LDM improved prostate cancer detection performance using synthetic MRI for classifier training but was limited to the axial T2-weighted (AxT2) sequence, did not investigate inter-institutional domain shift, and prioritized radiology over histopathology outcomes. We propose CCELLA++ to address these limitations and improve clinical utility. Methods: CCELLA++ expands CCELLA for simultaneous biparametric prostate MRI (bpMRI) generation, including the AxT2, high b-value diffusion series (HighB) and apparent diffusion coefficient map (ADC). Domain adaptation was investigated by pretraining classifiers on real or LDM-generated synthetic data from an internal institution, followed with fine-tuning on progressively smaller fractions of an out-of-distribution, external dataset. Results: CCELLA++ improved 3D FID for HighB and ADC but not AxT2 (0.013, 0.012, 0.063 respectively) sequences compared to CCELLA (0.060). Classifier pretraining with CCELLA++ bpMRI outperformed real bpMRI in AP and AUC for all domain adaptation scenarios. CCELLA++ pretraining achieved highest classifier performance below 50% (n=665) external dataset volume. Conclusion: Synthetic bpMRI generated by our method can improve downstream classifier generalization and performance beyond real bpMRI or CCELLA-generated AxT2-only images. Future work should seek to quantify medical image sample quality, balance multi-sequence LDM training, and condition the LDM with additional information. Significance: The proposed CCELLA++ LDM can generate synthetic bpMRI that outperforms real data for domain adaptation with a limited target institution dataset. Our code is available at https://github.com/grabkeem/CCELLA-plus-plus
Prompt-Guided Latent Diffusion with Predictive Class Conditioning for 3D Prostate MRI Generation
Jun 11, 2025Abstract:Latent diffusion models (LDM) could alleviate data scarcity challenges affecting machine learning development for medical imaging. However, medical LDM training typically relies on performance- or scientific accessibility-limiting strategies including a reliance on short-prompt text encoders, the reuse of non-medical LDMs, or a requirement for fine-tuning with large data volumes. We propose a Class-Conditioned Efficient Large Language model Adapter (CCELLA) to address these limitations. CCELLA is a novel dual-head conditioning approach that simultaneously conditions the LDM U-Net with non-medical large language model-encoded text features through cross-attention and with pathology classification through the timestep embedding. We also propose a joint loss function and a data-efficient LDM training framework. In combination, these strategies enable pathology-conditioned LDM training for high-quality medical image synthesis given limited data volume and human data annotation, improving LDM performance and scientific accessibility. Our method achieves a 3D FID score of 0.025 on a size-limited prostate MRI dataset, significantly outperforming a recent foundation model with FID 0.071. When training a classifier for prostate cancer prediction, adding synthetic images generated by our method to the training dataset improves classifier accuracy from 69% to 74%. Training a classifier solely on our method's synthetic images achieved comparable performance to training on real images alone.
A Modified AUC for Training Convolutional Neural Networks: Taking Confidence into Account
Jun 08, 2020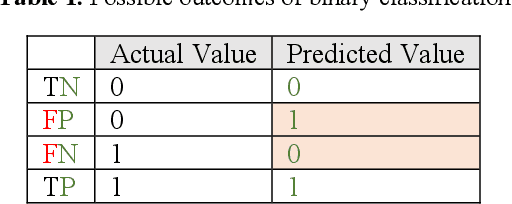
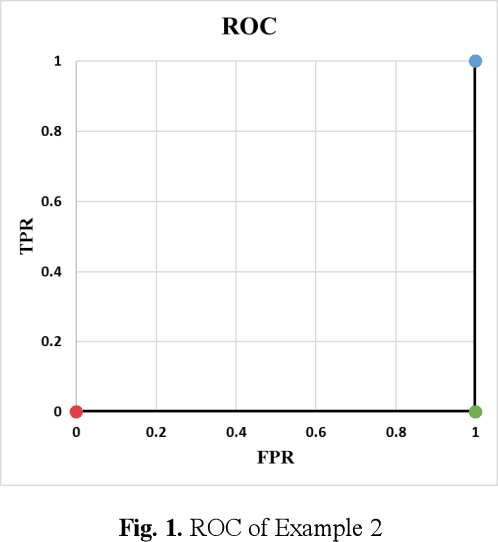
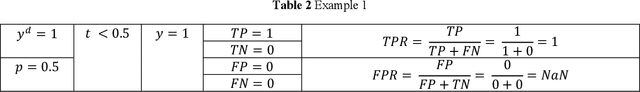
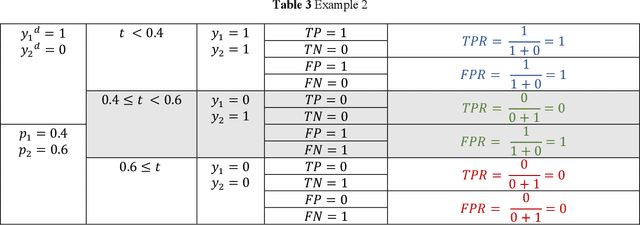
Abstract:Receiver operating characteristic (ROC) curve is an informative tool in binary classification and Area Under ROC Curve (AUC) is a popular metric for reporting performance of binary classifiers. In this paper, first we present a comprehensive review of ROC curve and AUC metric. Next, we propose a modified version of AUC that takes confidence of the model into account and at the same time, incorporates AUC into Binary Cross Entropy (BCE) loss used for training a Convolutional neural Network for classification tasks. We demonstrate this on two datasets: MNIST and prostate MRI. Furthermore, we have published GenuineAI, a new python library, which provides the functions for conventional AUC and the proposed modified AUC along with metrics including sensitivity, specificity, recall, precision, and F1 for each point of the ROC curve.
A Comprehensive Study of Data Augmentation Strategies for Prostate Cancer Detection in Diffusion-weighted MRI using Convolutional Neural Networks
Jun 01, 2020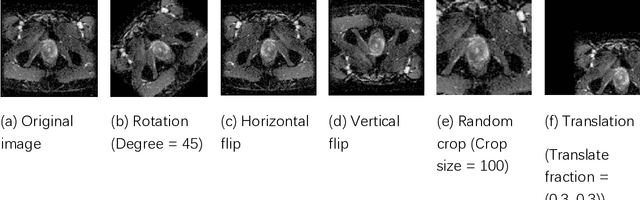
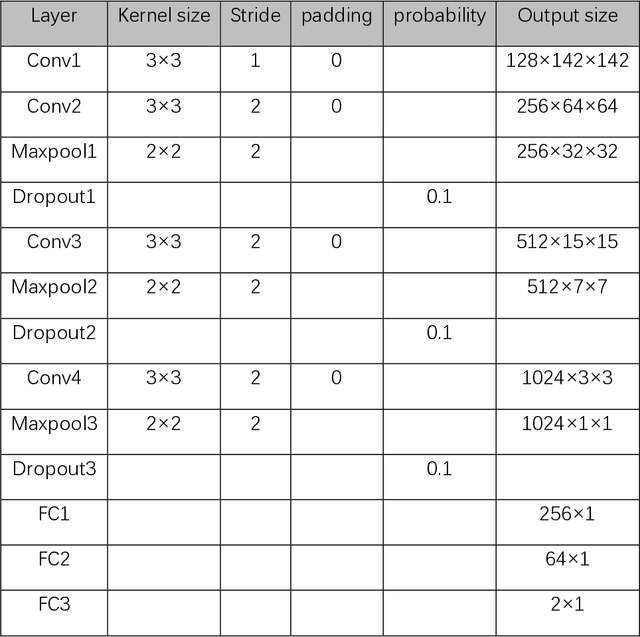
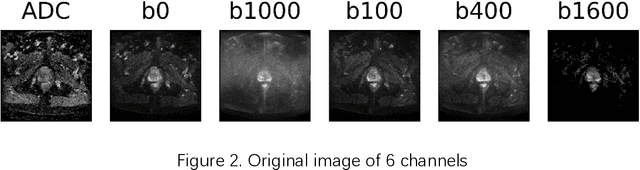
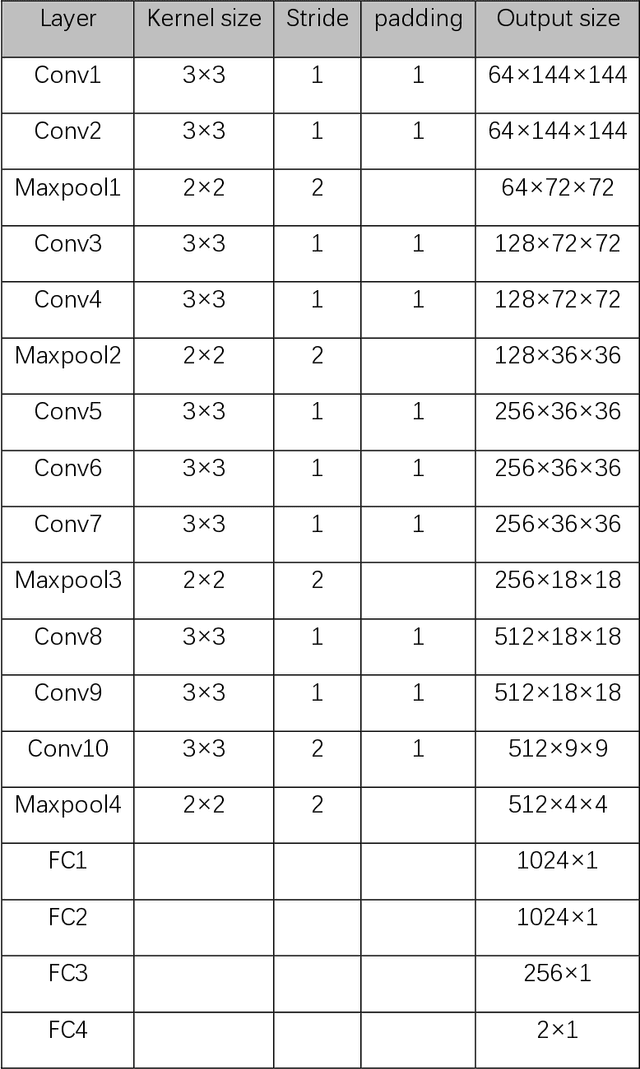
Abstract:Data augmentation refers to a group of techniques whose goal is to battle limited amount of available data to improve model generalization and push sample distribution toward the true distribution. While different augmentation strategies and their combinations have been investigated for various computer vision tasks in the context of deep learning, a specific work in the domain of medical imaging is rare and to the best of our knowledge, there has been no dedicated work on exploring the effects of various augmentation methods on the performance of deep learning models in prostate cancer detection. In this work, we have statically applied five most frequently used augmentation techniques (random rotation, horizontal flip, vertical flip, random crop, and translation) to prostate Diffusion-weighted Magnetic Resonance Imaging training dataset of 217 patients separately and evaluated the effect of each method on the accuracy of prostate cancer detection. The augmentation algorithms were applied independently to each data channel and a shallow as well as a deep Convolutional Neural Network (CNN) were trained on the five augmented sets separately. We used Area Under Receiver Operating Characteristic (ROC) curve (AUC) to evaluate the performance of the trained CNNs on a separate test set of 95 patients, using a validation set of 102 patients for finetuning. The shallow network outperformed the deep network with the best 2D slice-based AUC of 0.85 obtained by the rotation method.
Evolution-based Fine-tuning of CNNs for Prostate Cancer Detection
Nov 04, 2019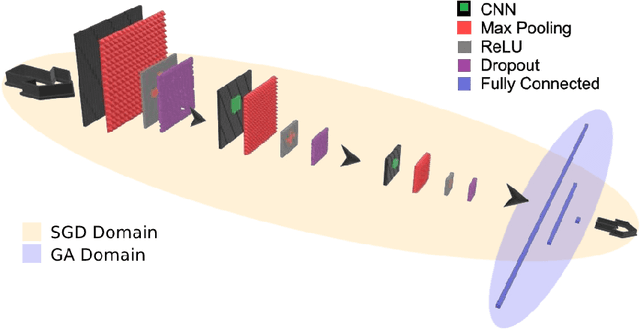
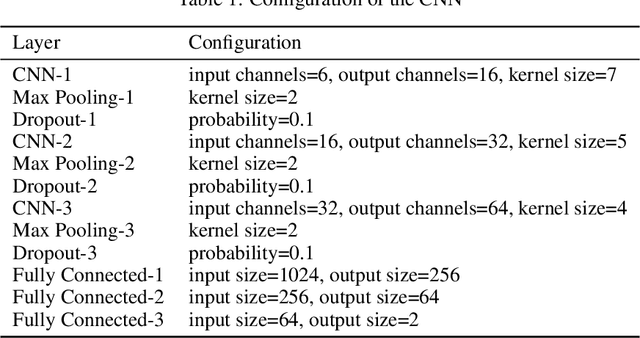
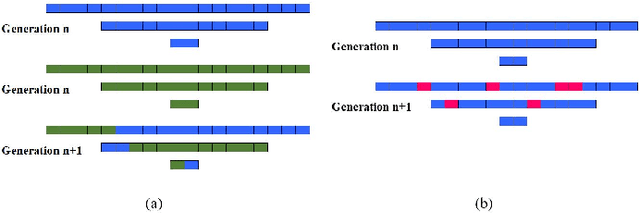
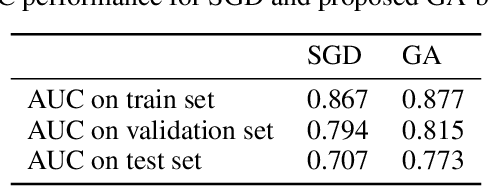
Abstract:Convolutional Neural Networks (CNNs) have been used for automated detection of prostate cancer where Area Under Receiver Operating Characteristic (ROC) curve (AUC) is usually used as the performance metric. Given that AUC is not differentiable, common practice is to train the CNN using a loss functions based on other performance metrics such as cross entropy and monitoring AUC to select the best model. In this work, we propose to fine-tune a trained CNN for prostate cancer detection using a Genetic Algorithm to achieve a higher AUC. Our dataset contained 6-channel Diffusion-Weighted MRI slices of prostate. On a cohort of 2,955 training, 1,417 validation, and 1,334 test slices, we reached test AUC of 0.773; a 9.3% improvement compared to the base CNN model.
A Transfer Learning Approach for Automated Segmentation of Prostate Whole Gland and Transition Zone in Diffusion Weighted MRI
Sep 20, 2019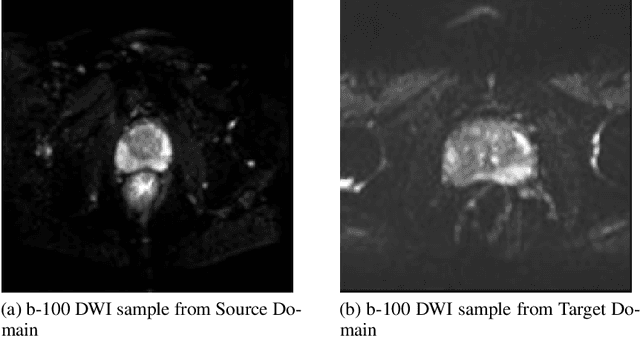
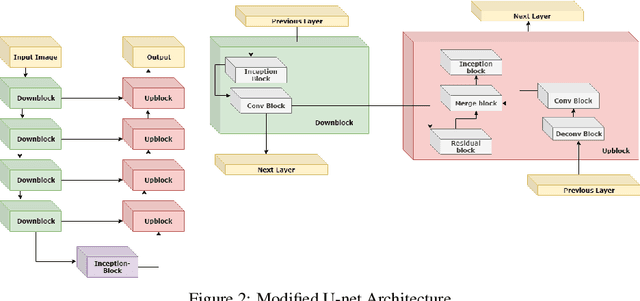
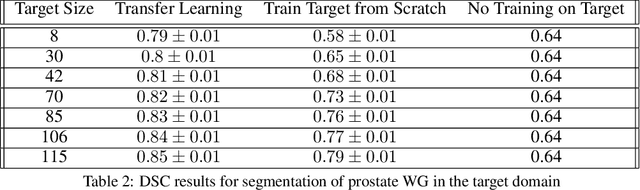
Abstract:The segmentation of prostate whole gland and transition zone in Diffusion Weighted MRI (DWI) are the first step in designing computer-aided detection algorithms for prostate cancer. However, variations in MRI acquisition parameters and scanner manufacturing result in different appearances of prostate tissue in the images. Convolutional neural networks (CNNs) which have shown to be successful in various medical image analysis tasks including segmentation are typically sensitive to the variations in imaging parameters. This sensitivity leads to poor segmentation performance of CNNs trained on a source cohort and tested on a target cohort from a different scanner and hence, it limits the applicability of CNNs for cross-cohort training and testing. Contouring prostate whole gland and transition zone in DWI images are time-consuming and expensive. Thus, it is important to enable CNNs pretrained on images of source domain, to segment images of target domain with minimum requirement for manual segmentation of images from the target domain. In this work, we propose a transfer learning method based on a modified U-net architecture and loss function, for segmentation of prostate whole gland and transition zone in DWIs using a CNN pretrained on a source dataset and tested on the target dataset. We explore the effect of the size of subset of target dataset used for fine-tuning the pre-trained CNN on the overall segmentation accuracy. Our results show that with a fine-tuning data as few as 30 patients from the target domain, the proposed transfer learning-based algorithm can reach dice score coefficient of 0.80 for both prostate whole gland and transition zone segmentation. Using a fine-tuning data of 115 patients from the target domain, dice score coefficient of 0.85 and 0.84 are achieved for segmentation of whole gland and transition zone, respectively, in the target domain.
Improving Prognostic Performance in Resectable Pancreatic Ductal Adenocarcinoma using Radiomics and Deep Learning Features Fusion in CT Images
Jul 10, 2019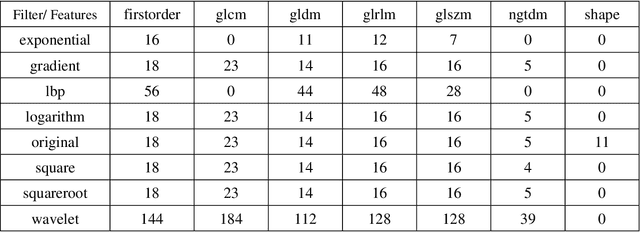
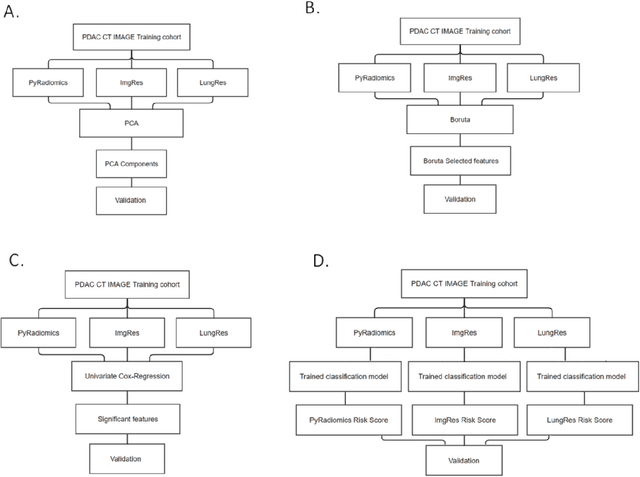
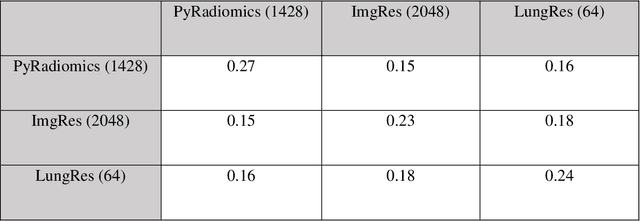
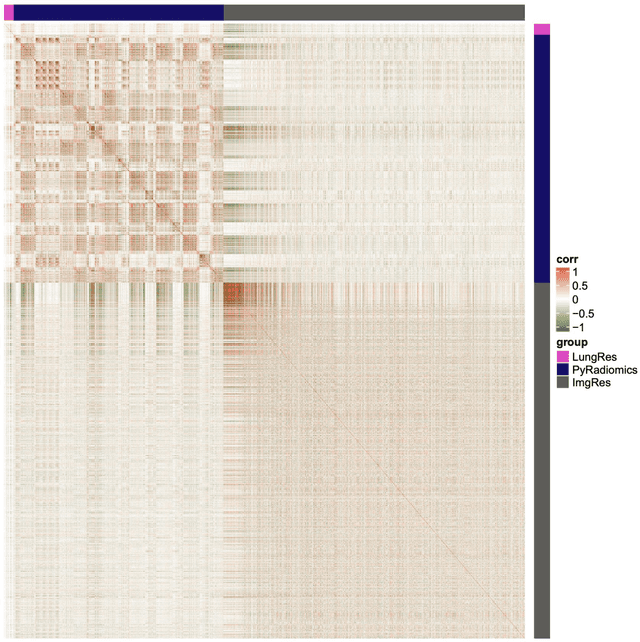
Abstract:As an analytic pipeline for quantitative imaging feature extraction and analysis, radiomics has grown rapidly in the past a few years. Recent studies in radiomics aim to investigate the relationship between tumors imaging features and clinical outcomes. Open source radiomics feature banks enable the extraction and analysis of thousands of predefined features. On the other hand, recent advances in deep learning have shown significant potential in the quantitative medical imaging field, raising the research question of whether predefined radiomics features have predictive information in addition to deep learning features. In this study, we propose a feature fusion method and investigate whether a combined feature bank of deep learning and predefined radiomics features can improve the prognostics performance. CT images from resectable Pancreatic Adenocarcinoma (PDAC) patients were used to compare the prognosis performance of common feature reduction and fusion methods and the proposed risk-score based feature fusion method for overall survival. It was shown that the proposed feature fusion method significantly improves the prognosis performance for overall survival in resectable PDAC cohorts, elevating the area under ROC curve by 51% compared to predefined radiomics features alone, by 16% compared to deep learning features alone, and by 32% compared to existing feature fusion and reduction methods for a combination of deep learning and predefined radiomics features.
CNN-based Survival Model for Pancreatic Ductal Adenocarcinoma in Medical Imaging
Jun 25, 2019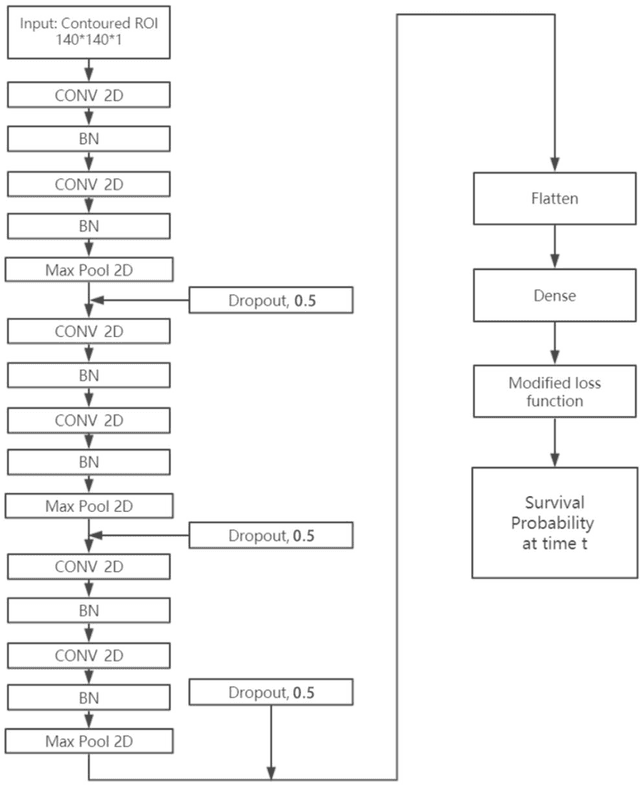
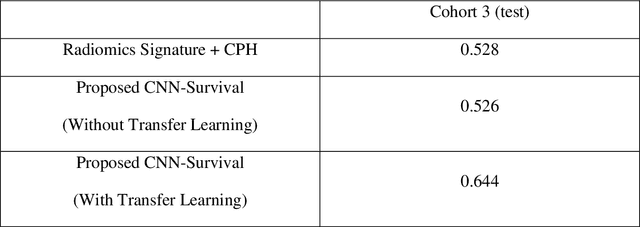
Abstract:Cox proportional hazard model (CPH) is commonly used in clinical research for survival analysis. In quantitative medical imaging (radiomics) studies, CPH plays an important role in feature reduction and modeling. However, the underlying linear assumption of CPH model limits the prognostic performance. In addition, the multicollinearity of radiomic features and multiple testing problem further impedes the CPH models performance. In this work, using transfer learning, a convolutional neural network (CNN) based survival model was built and tested on preoperative CT images of resectable Pancreatic Ductal Adenocarcinoma (PDAC) patients. The proposed CNN-based survival model outperformed the traditional CPH-based radiomics approach in terms of concordance index by 22%, providing a better fit for patients' survival patterns. The proposed CNN-based survival model outperforms CPH-based radiomics pipeline in PDAC prognosis. This approach offers a better fit for survival patterns based on CT images and overcomes the limitations of conventional survival models.
Prostate Cancer Detection using Deep Convolutional Neural Networks
May 30, 2019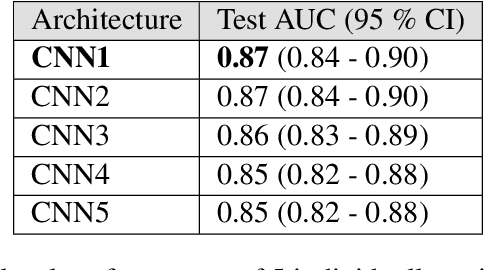
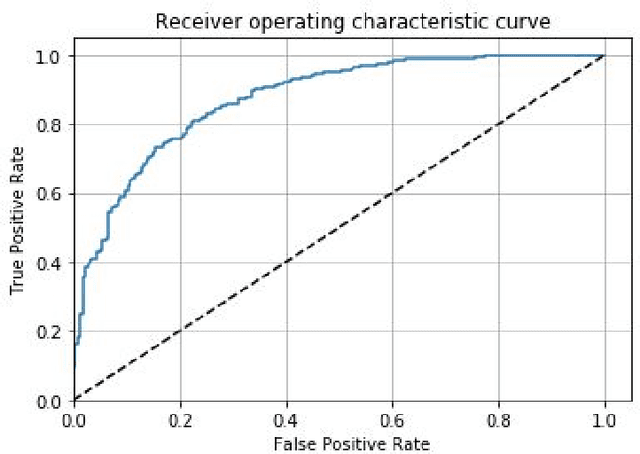
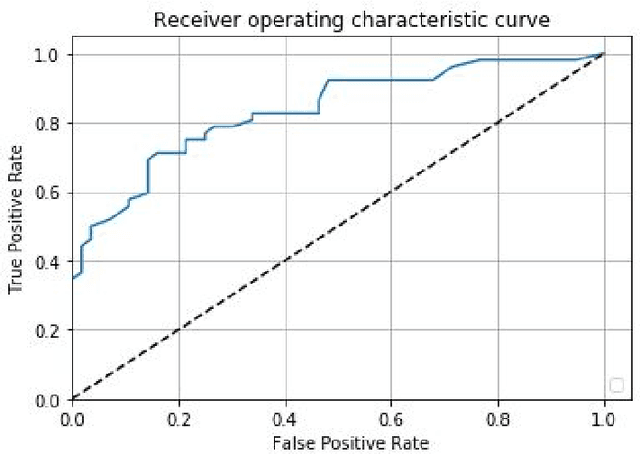
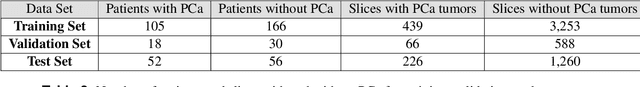
Abstract:Prostate cancer is one of the most common forms of cancer and the third leading cause of cancer death in North America. As an integrated part of computer-aided detection (CAD) tools, diffusion-weighted magnetic resonance imaging (DWI) has been intensively studied for accurate detection of prostate cancer. With deep convolutional neural networks (CNNs) significant success in computer vision tasks such as object detection and segmentation, different CNNs architectures are increasingly investigated in medical imaging research community as promising solutions for designing more accurate CAD tools for cancer detection. In this work, we developed and implemented an automated CNNs-based pipeline for detection of clinically significant prostate cancer (PCa) for a given axial DWI image and for each patient. DWI images of 427 patients were used as the dataset, which contained 175 patients with PCa and 252 healthy patients. To measure the performance of the proposed pipeline, a test set of 108 (out of 427) patients were set aside and not used in the training phase. The proposed pipeline achieved area under the receiver operating characteristic curve (AUC) of 0.87 (95% Confidence Interval (CI): 0.84-0.90) and 0.84 (95% CI: 0.76-0.91) at slice level and patient level, respectively.
Improving Prognostic Value of CT Deep Radiomic Features in Pancreatic Ductal Adenocarcinoma Using Transfer Learning
May 23, 2019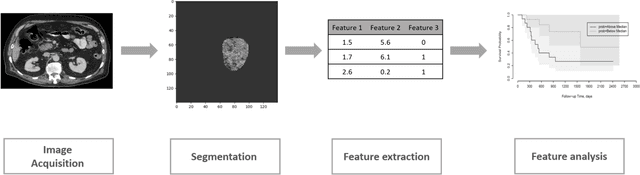
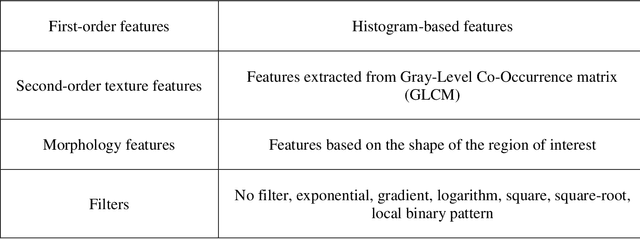
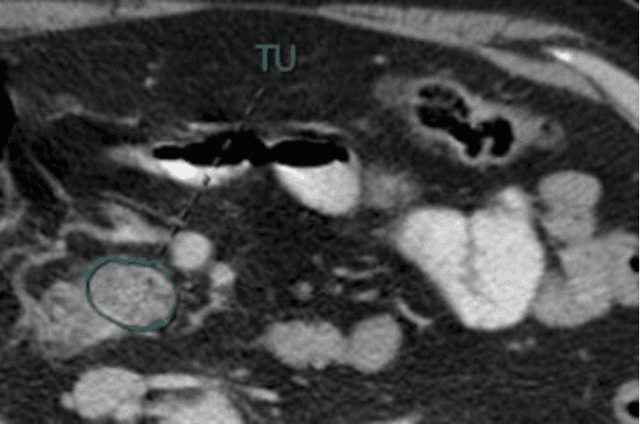
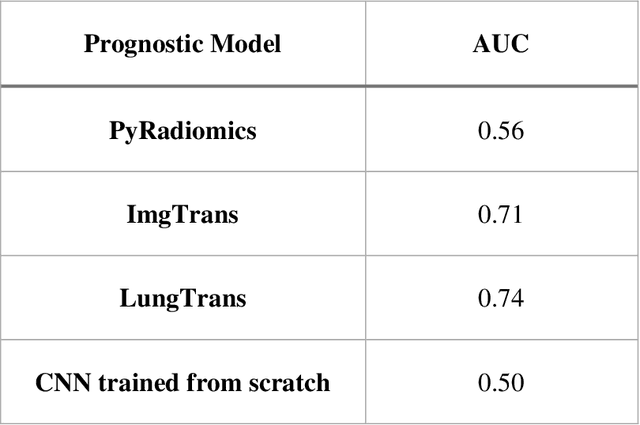
Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancers with an extremely poor prognosis. Radiomics has shown prognostic ability in multiple types of cancer including PDAC. However, the prognostic value of traditional radiomics pipelines, which are based on hand-crafted radiomic features alone, is limited due to multicollinearity of features and multiple testing problem, and limited performance of conventional machine learning classifiers. Deep learning architectures, such as convolutional neural networks (CNNs), have been shown to outperform traditional techniques in computer vision tasks, such as object detection. However, they require large sample sizes for training which limits their development. As an alternative solution, CNN-based transfer learning has shown the potential for achieving reasonable performance using datasets with small sample sizes. In this work, we developed a CNN-based transfer learning approach for prognostication in PDAC patients for overall survival. The results showed that transfer learning approach outperformed the traditional radiomics model on PDAC data. A transfer learning approach may fill the gap between radiomics and deep learning analytics for cancer prognosis and improve performance beyond what CNNs can achieve using small datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge